Unraveling the role of urea hydrolysis in salt stress response during seed germination and seedling growth in Arabidopsis thaliana
Figures
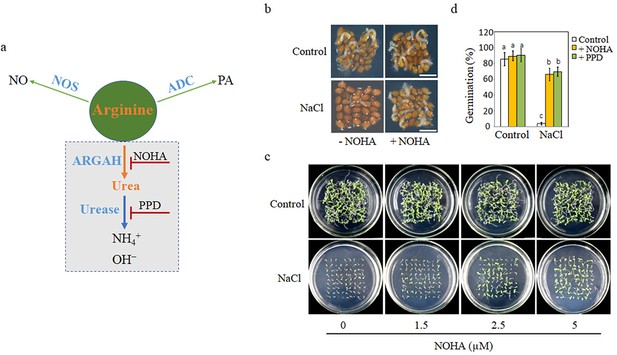
Blocking arginine hydrolysis and promoting seed germination under salt stress.
(a) A simple model of arginine metabolism in Arabidopsis thaliana. This model outlines the conversion of arginine into (1) nitric oxide (NO) and citrulline by nitric oxide synthase (NOS); (2) polyamine (PA) by arginine decarboxylase (ADC); and (3) ornithine and urea by arginase, with urea further decomposed to ammonia by urease. It highlights arginine as a shared competitive substrate for the three enzymes ARGAH, NOS, and ADC, illustrating the competitive enzymatic interactions. NOHA: NG-hydroxy-l-arginine, an arginase inhibitor; PPD: phenyl phosphorodiamidate, a urease inhibitor. (b) Comparison of germination rates of WT seeds on half-strength MS (½ MS) medium containing 0 mM (control) or 135 mM NaCl with 5 μM NOHA or without NOHA. Photographs were taken at the 48 hr after 2 days at 4°C. A representative result from one of three independent experiments, all yielding similar outcomes, is shown. Scale bar represents 1 mm. (c) Seeds were germinated on ½ MS medium containing either 0 mM or 135 mM NaCl and varying concentrations of NOHA (0, 1.5, 2.5, 5 μM); photographs were taken 14 days after germination. (d) Germination rates of WT seeds in either 0 mM or 135 mM NaCl medium with or without 5 μM NOHA or 15 μM PPD. Stratification consisted of pretreatment of seeds for 2 days at 4°C in the darkness. In all experiments, seeds were freshly sowed and incubated under 16 hr light and 8 hr dark conditions at 22℃. The experiment was repeated three times; at least 30 seeds were counted in each replicate. The data was analyzed using a one-way ANOVA, followed by Duncan’s multiple range test for the post hoc comparisons. Significant differences between groups, indicated by different letters on the error bars, were determined (p<0.05).

Effects of NG-hydroxy-l-arginine (NOHA) on WT seedling growth.
(a, b) Growth of WT seedlings, on ½ MS without (a) and with 135 mM NaCl (b) both subjected to treatments of 0, 1.5, 2.5, and 5 µM NOHA. Photographs captured 14 days after germination. (c, d) Root length was measured across both control and salt-stressed seedlings. In all experiments, seeds were freshly sowed seeds and incubated under a 16 hr light/8 hr dark condition cycle at 22℃. The experiment was repeated three times with at least 35 seeds per replicate. The data was analyzed using a one-way ANOVA and Duncan’s post hoc test. Different letters on the error bars represent significant differences (p<0.05).
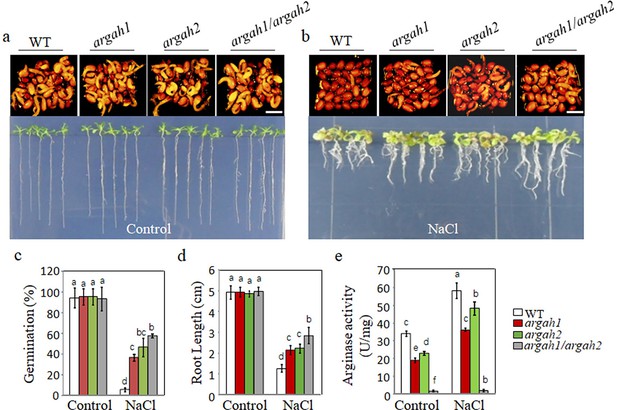
Deletion of arginase hydrolytic pathway alleviates the inhibition of seed germination by salt.
WT, argah1, argah2, and argah1/argah2 mutant seeds germinated, and seedling growth was observed on ½ MS medium under two conditions: 0 mM NaCl (control) (a) and 135 mM NaCl (b). Seedlings images of seeds germination 48 hr after 2 days at 4°C and 14 days after germination. (c) Germination rates, (d) root length measurements, and (e) arginase activity assays for WT, argah1, argah2, and argah1/argah2 mutants on control and 135 mM NaCl mediums. In all experiments, seeds were freshly sowed and incubated under 16 hr light and 8 hr dark conditions at 22℃. The experiment was repeated three times; at least 30 seeds were counted in each replicate. Scale bar: 1 mm. Data were subjected to a one-way ANOVA, followed by Duncan’s post hoc test. Different letters on the error bars denote significant differences in the data (p<0.05).

Characterization of atargah1, atargah2, and the atargah1/atargah2 double mutant plants.
(a) Characterization of atargah1 and atargah2, an Arabidopsis T-DNA insertion mutant, green boxes represent exons, and black lines represent introns. (b) AtArgAH1 and AtArgAH2 double genes knockout mediated by the CRISPR-Cas9 system; schematic map of the sgRNA targeted sites on the genomic regions of AtArgAH1 and AtArgAH2, and sequencing results of the AtArgAH1/AtArgAH2 double homozygous mutant lines atargah1/atargah2 from the T1 generation. The edited exon is shown as green boxes, the adjacent intron is shown as lines, and black arrows represent insertions.
-
Figure 2—figure supplement 1—source data 1
Sequencing results of the AtArgAH1/AtArgAH2 double homozygous mutant lines atargah1/atargah2.
Genomic DNA was extracted from young leaves of WT and transformed plants, and amplified by PCR using primers flanking the target sites to confirm the introduction of mutations. The PCR products were sequenced to identify double mutants, atargah1/atargah2. The primer sequences were using primers AtArgAH1-FW/RV and AtArgAH2-FW/RV (Supplementary file 1). Inset bases in the atargah1/atargah2 mutant sequence are highlighted in yellow.
- https://cdn.elifesciences.org/articles/96797/elife-96797-fig2-figsupp1-data1-v3.docx
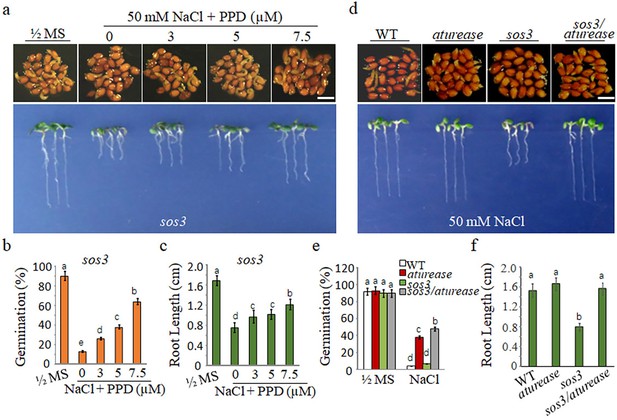
Blocking the arginine hydrolysis pathway mitigates the hypersensitivity of sos3 mutants to salt.
(a) Seeds germinated and seedling growth of sos3 mutants on ½ MS with 50 mM NaCl medium, supplemented with phenyl phosphorodiamidate (PPD) at different concentrations (0, 3, 5, and 7.5 μM). Seedlings images of seeds germination 48 hr after 2 days at 4°C and 14 days after germination. (b) Germination rates and (c) root lengths of sos3 mutants assessed after the specified treatments. (d) WT, aturease, sos3, and sos3/aturease seeds germinated and grew on ½ MS with 50 mM NaCl. Seedlings images of seeds germination 48 hr after 2 days at 4°C and 14 days after germination. (e) Germination rate and (f) root lengths for WT, sos3, aturease, and sos3/aturease mutants were measured after the indicated treatments. In all experiments, seeds were freshly sowed and incubated under a 16 hr light/8 hr dark cycle at 22℃. The experiment was repeated three times; at least 25 seeds were counted in each replicate. Scale bar: 1 mm. The data were analyzed using one-way ANOVA followed by Duncan’s post hoc test, with different letters on the error bars indicating significant differences in the data (p<0.05).
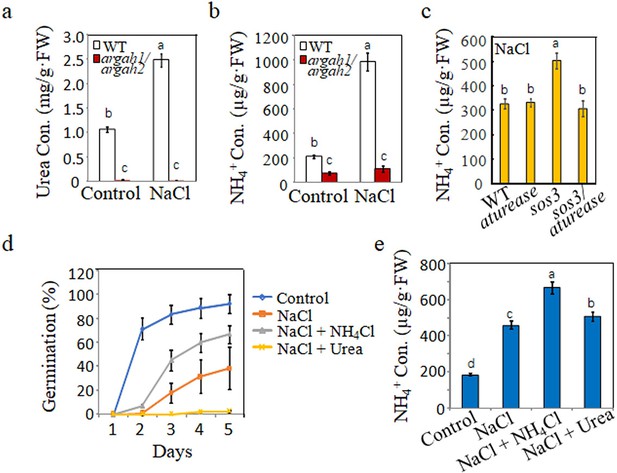
Correlation between metabolites from arginine hydrolysis and salt inhibit-induced inhibition of seed germination.
(a) Urea and (b) NH4+ concentrations were measured in WT and argah1/argah2 seedlings grown in ½ MS medium under control (0 mM) and 135 mM NaCl conditions. (c) NH4+ levels assessed in WT, aturease, sos3, and sos3/aturease seedlings under 50 mM NaCl treatment in ½ MS. (d) Effect of NH4Cl on salt response was evaluated in WT seeds, monitoring germination rates in control medium with 135 mM NaCl, with and without 10 mM NH4Cl and 10 mM urea, over time. (e) NH4+ concentration in WT seedlings measured on ½ MS with 135 mM NaCl, with or without the addition of 10 mM NH4Cl and 10 mM urea. In all experiments, seeds were freshly sowed and incubated under a 16 hr light/8 hr dark cycle at 22℃. The experiment was repeated three times; at least 30 seeds were counted in each replicate. Scale bar: 1 mm. The data were analyzed using a one-way ANOVA, with Duncan’s test for post hoc comparison. Different letters on the error bars indicate significant differences in the data (p<0.05).

Effect of NH4+ or urea on NaCl responses of wild-type (WT) germinated seeds.
(a) Growth of WT seedlings on ½ MS medium (control; without 135 mM NaCl or 10 mM NH4Cl, NH4NO3, urea), on ½ MS medium with 135 mM NaCl, and on ½ MS medium with 135 mM NaCl containing 10 mM NH4Cl, or NH4NO3, or urea. Photographs were taken 14 days after germination. (b) Root length was measured after the indicated treatments. In all experiments, seeds were freshly sowed and incubated under 16 hr light and 8 hr dark conditions at 22℃. The experiment was repeated three times with at least 30 seeds per replicate. The data analysis was performed using a one-way ANOVA and Duncan’s post hoc test, with different letters denoting significant differences (p<0.05).
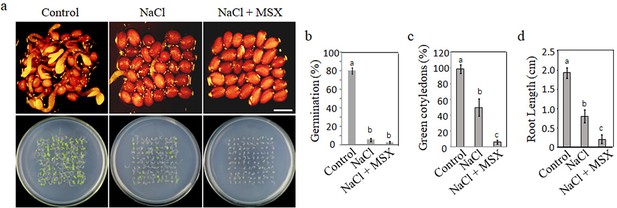
Effect of acidification process of glutamine synthetase assimilation on plant salt tolerance.
(a) Germination of WT seeds on ½ MS medium (control), ½ MS medium supplemented with 135 mM NaCl, and ½ MS medium containing 3 µM l-methionine sulfoximine (MSX). Seedlings images of seeds germination 48 hr after 2 days at 4°C and 10 days after germination. Scale bar represents 1 mm. The study evaluated (b) WT seeds germination rates, (c) the count of green cotyledons, and (d) root length. In all experiments, seeds were freshly sowed and incubated under 16 hr light and 8 hr dark conditions at 22℃. The experiment was repeated in triplicate with a minimum of 30 seeds per replicate. The data analysis was performed using a one-way ANOVA and Duncan’s post hoc test, with significant differences (p<0.05) denoted by different letters on the error bars.
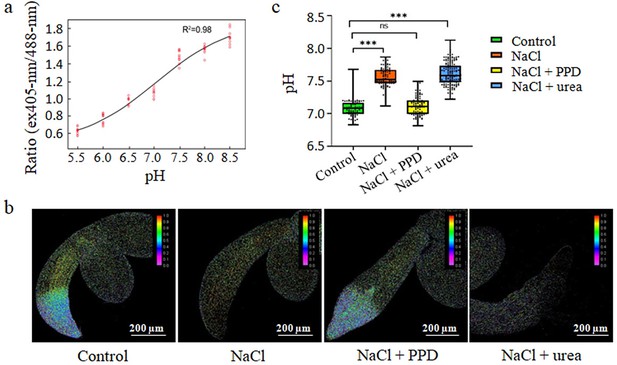
Inhibition of urea via arginine hydrolysis promotes seed germination under salt stress by lowering cell pH.
(a) Calibration curve for pHluorin in seedlings adjusted to various pH levels using 50 mM MES-BTP (pH 5.2–6.4), 50 mM HEPES-BTP (pH 6.8–7.6), and 50 mM ammonium acetate. The curve plots the average fluorescence intensity ratios against the pH for 15 seedlings. (b) Fluorescence ratio images (emission 500–550 nm) of PRpHluorin expression seedlings grown in control, 135 mM NaCl medium, 135 mM NaCl with 15 µM PPD, and 135 mM NaCl with 10 mM urea, at 22℃ for 3 days. Scale bar represents 200 μm. (c) Boxplots depict the cytoplasmic pH in root epidermal cells of seedlings, measured using the PRpHluorin fluorescence in the root elongation zone under treatments. Middle horizontal bars of boxplots represent the median, the bottom and top represent the 25th and 75th percentiles, and whiskers extend to the minimum and maximum. Statistical significance (***p<0.001) was revealed by the Student’s t-test. The experiment was repeated more than three times with similar results.

Cytoplasmic pH of the root epidermal cells measurement.
For pH calculation after each treatment, 10 PRpHluorin-expressing seedlings, with 100 cells per root, were analyzed. The epidermal cells of the root elongation zone were selected for pH measurements. The selected area is represented by the yellow dotted boxes, and pH values were extrapolated from the sigmoidal function established in vitro calibration curves using ImageJ. Statistical analysis was conducted using Student’s t-test. For visual representation, the grayscale ratio images were converted into pseudocolored images using the ImageJ illustrating the pH profiles.
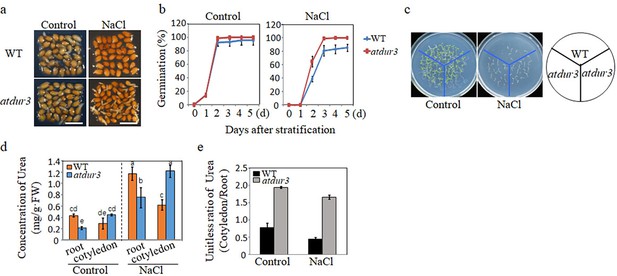
Arginine-derived urea transport and its role in salt-inhibited seed germination (SISG).
(a) Seed germination phenotypes of WT and atdur3 mutants were defined as the initial emergence of the radicle, which was observed and recorded at 48 hr of incubation following a 2-day stratification period at 4°C. Photographs captured under a stereomicroscope. Scale bar = 1 mm. (b) Germination rates of atdur3 on ½ MS medium, containing 0 (control) or 135 mM NaCl, respectively, over time. (c) Growth comparison of WT and atdur3 mutant seedlings on control and NaCl medium, photographed 10 days after germination. (d) Urea content analysis in roots and cotyledons of 5-day-old seedlings grown on control or 135 mM NaCl medium. (e) Calculation of urea (cotyledon/root) change ratio. In all experiments, seeds were freshly sowed and incubated under 16 hr light and 8 hr dark conditions at 22℃. The experiment was repeated three times; at least 30 seeds were counted in each replicate. The data was analyzed using a one-way ANOVA and Duncan’s post hoc test, with different letters indicating significant differences (p<0.05).

Characterization of atdur3 mutant plants.
(a) T-DNA insertion site in Dur3; gray boxes indicate exons; black lines indicate introns. (b) RT-PCR analysis of AtDur3 expression in the WT and atdur3 mutants. Actin expression was used as an internal control for normalization of the RT-PCR expression data.
-
Figure 7—figure supplement 1—source data 1
Complete original file of the full raw unedited AtDur3 RT-PCR gel image.
The AtDur3 gene was amplified using the specific primers AtDur3-FW and AtDur3-RV.
- https://cdn.elifesciences.org/articles/96797/elife-96797-fig7-figsupp1-data1-v3.zip
-
Figure 7—figure supplement 1—source data 2
Complete original file of the full raw unedited Actin gel image.
The Actin gene was amplified using the primers AtActin-FW and AtActin-RV.
- https://cdn.elifesciences.org/articles/96797/elife-96797-fig7-figsupp1-data2-v3.zip
-
Figure 7—figure supplement 1—source data 3
RT-PCR expression data of AtDur3 in the WT and atdur3 mutants.
Total RNA was extracted from young leaves of WT and atdur3 plants. Gene-specific primers pairs AtDur3-FW and AtDur3-RV were used for AtDur3, while AtActin-FW and AtActin-RV were used for Actin (Supplementary file 1). Actin expression was used as an internal control for normalization of the RT-PCR expression data.
- https://cdn.elifesciences.org/articles/96797/elife-96797-fig7-figsupp1-data3-v3.docx
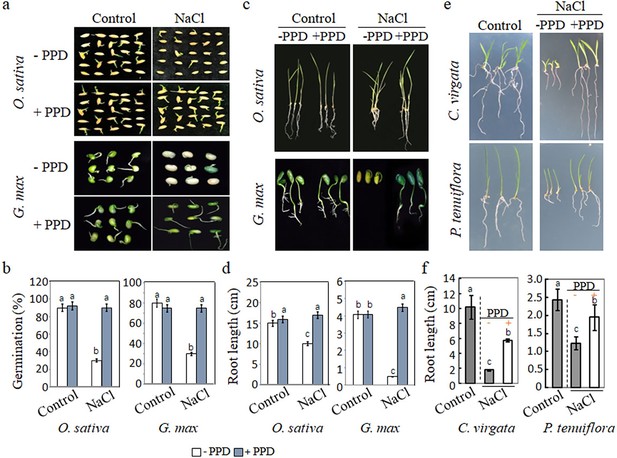
Effect of urea hydrolase inhibitor phenyl phosphorodiamidate (PPD) on the germination and seedling growth of Oryza sativa and Glycine max, Chloris virgata and Puccinellia tenuiflora.
(a) Seeds of O. sativa and G. max were germinated on ½ MS medium (control; 0 mM NaCl) or ½ MS containing 150 mM NaCl with or without 15 µM PPD. Photographs were taken 14 days after germination. (b) Germination rates of O. sativa and G. max plants in control or 150 mM NaCl with or without 15 µM PPD were calculated after 5-day sowing. (c) Seedling growth of O. sativa and G. max treated with and without PPD under salt stress, photographed 14 days after germination. (d) Root lengths were determined 14 days after germination. (e) Seeds of C. virgata and P. tenuiflora were germinated 150 mM NaCl with or without 15 µM PPD compared to control (0 mM NaCl). Representative morphological images of the treated seedlings 14 days after germination. (f) Root lengths were determined 14 days after germination. The experiment was repeated three times with at least 30 plants per replicate. PPD, an inhibitor of urea hydrolase and a downstream metabolite of arginine.
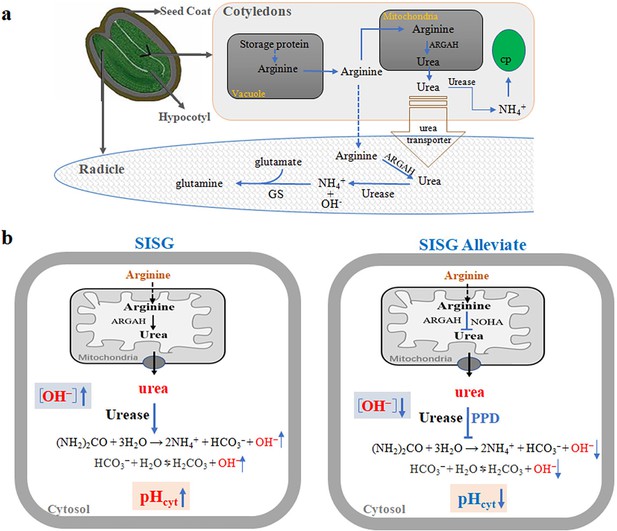
Hypothetical model for the regulation of seed germination by the arginine hydrolysis pathway under salt stress condition.
(a) Salt stress highly induced the accumulation of arginine through the degradation of seed-stored proteins in the cotyledon. Then, arginine is hydrolyzed to urea through the action of arginine amidinohydrolase (ARGAH), and urea is further degraded by plant urease to form NH4+ and OH-. (b) Salt stress stimulates the arginine hydrolysis pathway, leading to urea production by ARGAH, and its subsequent breakdown by urease, increasing NH4+, HCO3-, OH- levels, and cytoplasmic pH (pHcyt), which inhibits seed germination (SISG). Application of arginine hydrolysis inhibitors (e.g., NG-hydroxy-l-arginine [NOHA] for arginase, and phenyl phosphorodiamidate [PPD] for urease) or using ARGAH or Urease gene deletion mutants under salt stress can reduce NH4+, HCO3-, OH- levels, and decreased pHcyt, thus promoting seed germination (SISG alleviate). The model delineates the urea hydrolysis process, resulting in increased cytoplasmic pH, NH4+, and alkalinity, contributing to the inhibition of seed germination under salt conditions, abbreviated as SISG (salt inhibits seed germination). PPD, an inhibitor of urea hydrolase.

Effect of AtUrease inhibitor PPD on growth of Wild-type (WT) and atdur3.(A-D) germinated on half-strength MS or 135 mM NaCl with or without 15 µM PPD.
Representative images show the morphology of seedlings 14 d after 2 days stratification at 4℃. (E) Root length was determined 14 d after the stratification.
Tables
| Reagent type (species) or resource | Designation | Source or reference | Identifiers | Additional information |
|---|---|---|---|---|
| Genetic reagent (Arabidopsis thaliana) | Arabidopsis thaliana mutant (atargah1) | ABRC (Ohio State University) | SALK_057987 | |
| Recombinant DNA reagent | CR-PCR-AtArgAH1AtArgAH2 (plasmid) | This paper | M2CRISPR vector | |
| Sequence-based reagent | atargah1/atargah2 | This paper | PCR primers AtArgAH1- FW/ RV | ACATGGGTTTCATTATGAAC/ CACAAAAGACTAAATACATG |
| Sequence-based reagent | atargah1/atargah2 | This paper | PCR primers AtArgAH2- FW/ RV | CCTTGCGGTCCTTGCCAAC/ ATAAACAGAATCTTATTGAG |
| Commercial assay or kit | Arginase Assay Kit | Bioassay | DARG-048 | |
| Commercial assay or kit | Urea Assay Kit | Bioassay | DIUR-048 | |
| Chemical compound, drug | NG-Hydroxy-l-arginine | Sigma-Aldrich | H7278 | |
| Chemical compound, drug | Phenyl phosphorodiamidate | Macklin | p858176 | |
| Chemical compound, drug | l-Methionine sulfoximine | Sigma-Aldrich | M5379 | |
| Software, algorithm | SPSS | SPSS | RRID:SCR_002865 |
Additional files
-
Supplementary file 1
PCR primer sequences used in the current study.
The T-DNA insertion homozygous lines SALK_057987 (atargah1) were amplified using primers 057987-LP or 057987-RP and a T-DNA primer LBP. The AtDur3 gene was amplified using the specific primers AtDur3-FW and AtDur3-RV. The Actin gene was amplified using the primers AtActin-FW and AtActin-RV. The PCR products of double mutants atargah1/atargah2 were sequenced using primers AtArgAH1-FW/RV and AtArgAH2-FW/RV. The PRpHluorin gene was amplified using the specific primers PRpHluorin-F and PRpHluorin-R.
- https://cdn.elifesciences.org/articles/96797/elife-96797-supp1-v3.docx
-
MDAR checklist
- https://cdn.elifesciences.org/articles/96797/elife-96797-mdarchecklist1-v3.docx





