The Arthropoda-specific Tramtrack group BTB protein domains use previously unknown interface to form hexamers
Figures
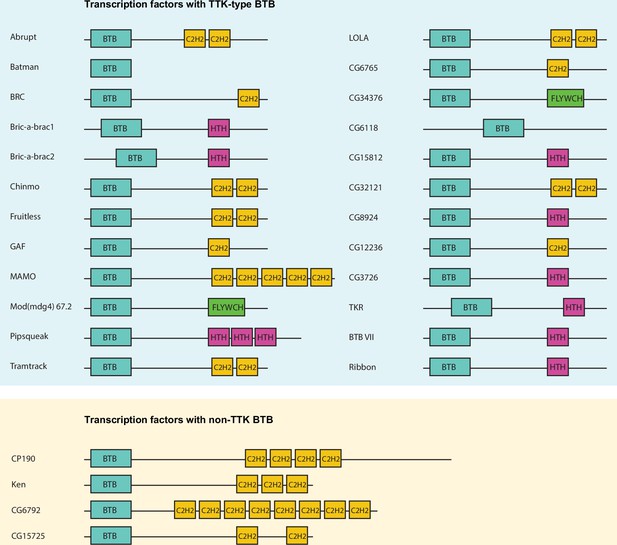
Schematic representation of Drosophila transcription factors with broad-complex, Tramtrack, and bric-a-brac (BTB) domains.
Only one representative isoform is shown for each gene. Pipsqueak psq DNA-binding domain is depicted as ‘HTH’ since it is a type of helix-turn-helix (HTH) domain.
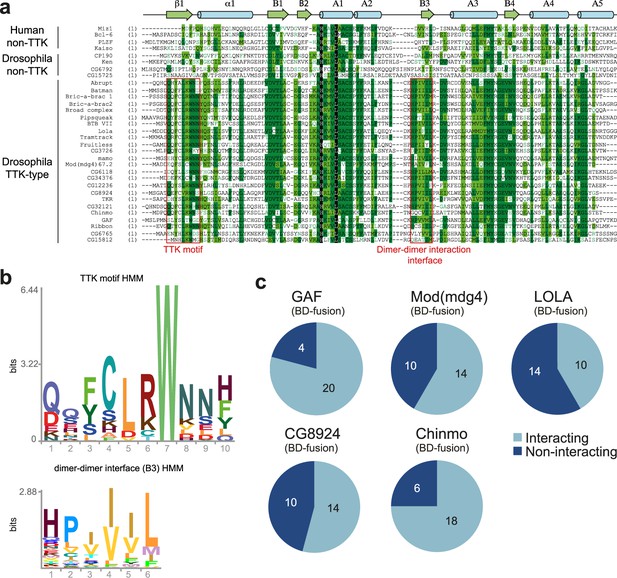
Characterization of the the Tramtrack group (TTK)-type broad-complex, Tramtrack, and bric-a-brac (BTB) domains.
(a) Multiple sequence alignment of BTB domains from Drosophila transcription factors and a few human BTB domains with known 3D structures. Secondary structure elements are labeled according to Stogios et al., 2005.(b) Hidden Markov Model (HMM)-profile models for the TTK motif (upper) and the main dimer-dimer interaction interface (lower) were obtained for 14 Diptera species. (c) Testing of the GAF, Mod(mdg4), and LOLA BTB domains for interaction with all TTK-type BTB domains found in Drosophila melanogaster. Original data are shown in Figure 2—figure supplement 1.
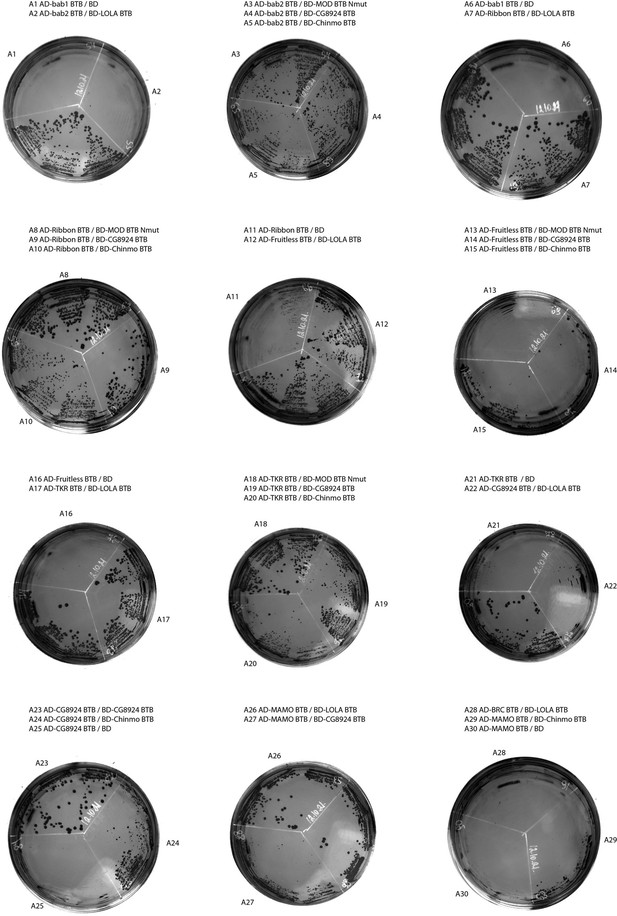
Results of yeast two-hybrid assays.
Growth assay plates without histidine are shown (yeasts are unable to grow on this medium in the absence of interaction). AD stands for Activation Domain, BD – for DNA-Binding Domain of GAL4 protein. Unlabeled plate segments correspond to excessive controls.
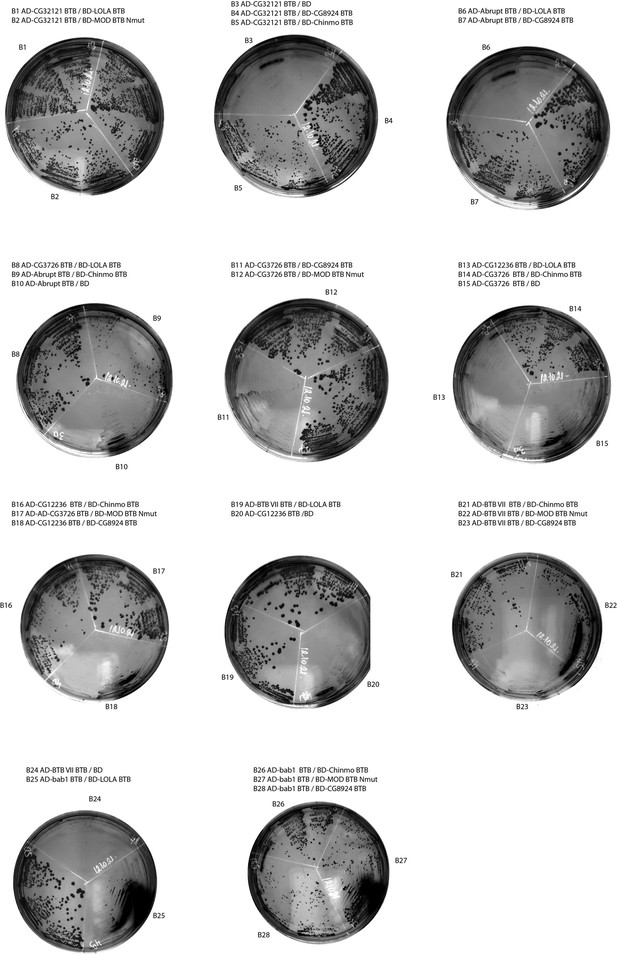
Results of yeast two-hybrid assays of interactions between the Tramtrack group (TTK) broad-complex, tramtrack, and bric-a-brac (BTB) domains (continued).
Designations are the same as in Figure 2—figure supplement 1.
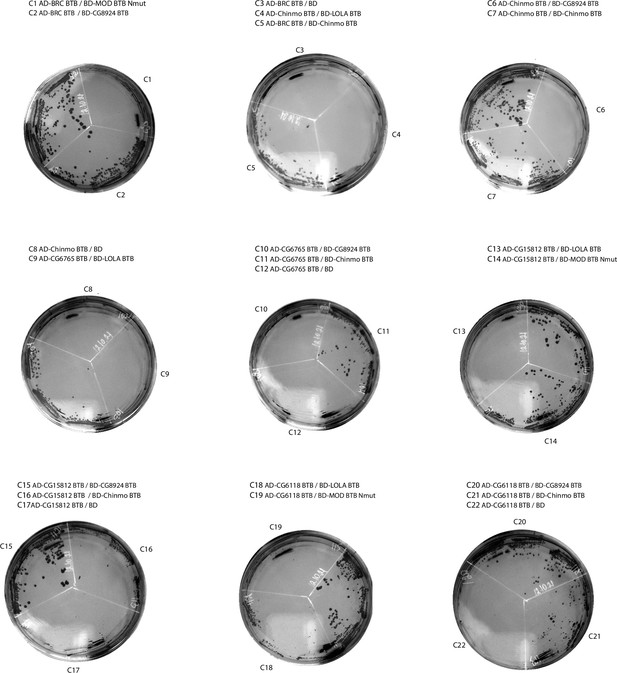
Results of yeast two-hybrid assays of interactions between The Tramtrack group (TTK) Broad-complex, tramtrack, and bric-a-brac (BTB) domains (continued).
Designations are the same as in Figure 2—figure supplement 1.
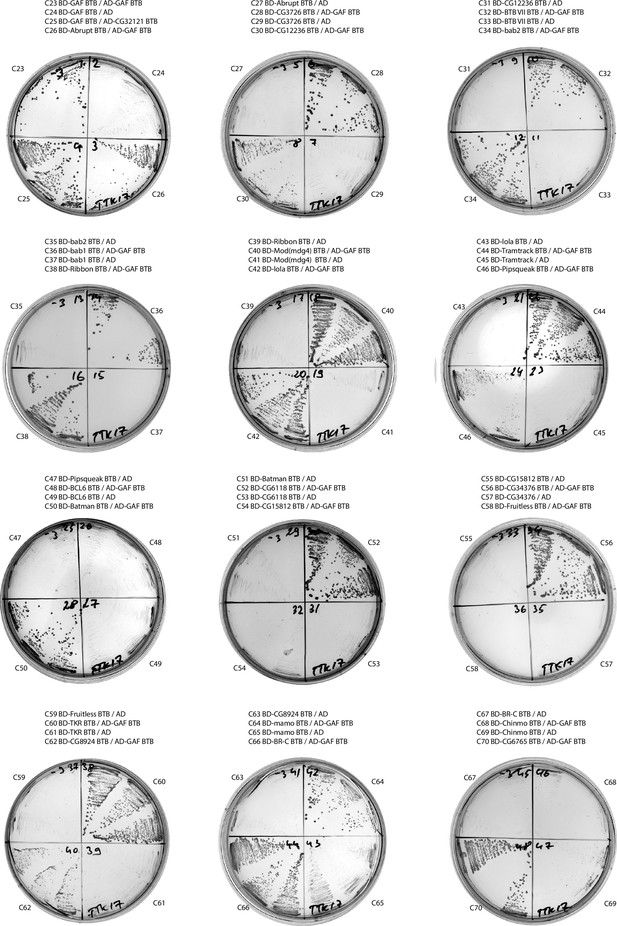
Results of yeast two-hybrid assays of interactions between the Tramtrack group (TTK) broad-complex, tramtrack, and bric-a-brac (BTB) domains (continued).
Designations are the same as in Figure 2—figure supplement 1.

Results of yeast two-hybrid assays of interactions between the Tramtrack group (TTK) broad-complex, tramtrack, and bric-a-brac (BTB) domains (continued).
Designations are the same as in Figure 2—figure supplement 1.
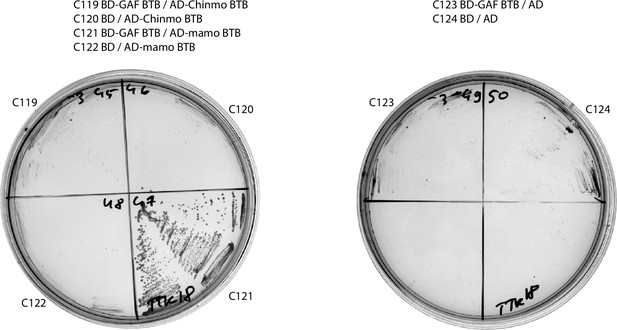
Results of yeast two-hybrid assays of interactions between the Tramtrack group (TTK) broad-complex, tramtrack, and bric-a-brac (BTB) domains (continued).
Designations are the same as in Figure 2—figure supplement 1.
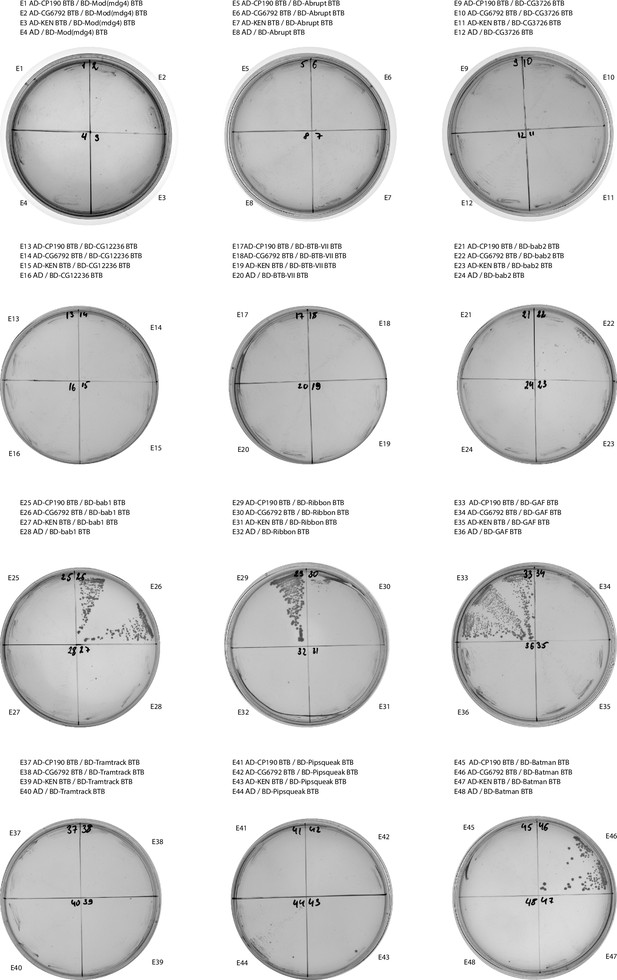
Results of yeast two-hybrid assays of interactions between the Tramtrack group (TTK) and non-TTK broad-complex, tramtrack, and bric-a-brac (BTB) domains.
Designations are the same as in Figure 2—figure supplement 1.
Results of yeast two-hybrid assays of interactions between the Tramtrack group (TTK) and non-TTK broad-complex, tramtrack, and bric-a-brac (BTB) domains (continued).
Designations are the same as in Figure 2—figure supplement 1.
Results of yeast two-hybrid assays of interactions between the Tramtrack group (TTK) and non-TTK broad-complex, tramtrack, and bric-a-brac (BTB) domains (continued).
Designations are the same as in Figure 2—figure supplement 1.
Results of yeast two-hybrid assays of interactions between the Tramtrack group (TTK) and non-TTK broad-complex, tramtrack, and bric-a-brac (BTB) domains (continued).
Designations are the same as in Figure 2—figure supplement 1.
Results of yeast two-hybrid assays of interactions between the Tramtrack group (TTK) and non-TTK broad-complex, tramtrack, and bric-a-brac (BTB) domains (continued).
Designations are the same as in Figure 2—figure supplement 1.
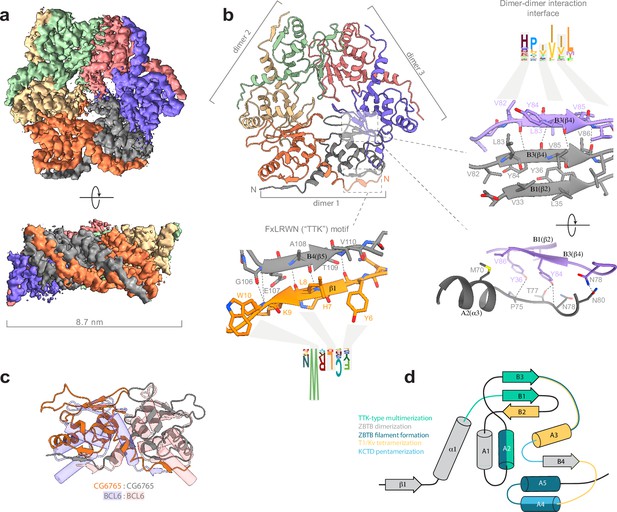
Cryo-EM structure of CG6765 broad-complex, tramtrack, and bric-a-brac (BTB) domain.
(a) Cryo-EM map of CG6765 BTB domain. Map regions are colored according to corresponding protein chains. (b) Refined model of CG6765 BTB domain hexamer. Individual dimers are depicted, and details of dimer-dimer interaction interface and beta-sheet formation by FxLRWN (‘the Tramtrack group, TTK’) motif are shown at the right. Secondary structure elements are depicted according to Stogios et al., 2005. (c) Overlay of the dimeric subunit of CG6765 BTB domain and classical dimer of Bcl6 BTB domain (PDB ID: 1R28) (Ahmad et al., 2003). (d) Summary of the involvement of the secondary structural elements of BTB domains in multimerization. Overlays of BTB multimeric assemblies are shown at the Figure 3—figure supplement 5. (d) has been adapted from Figure 3A from Bonchuk et al., 2023.
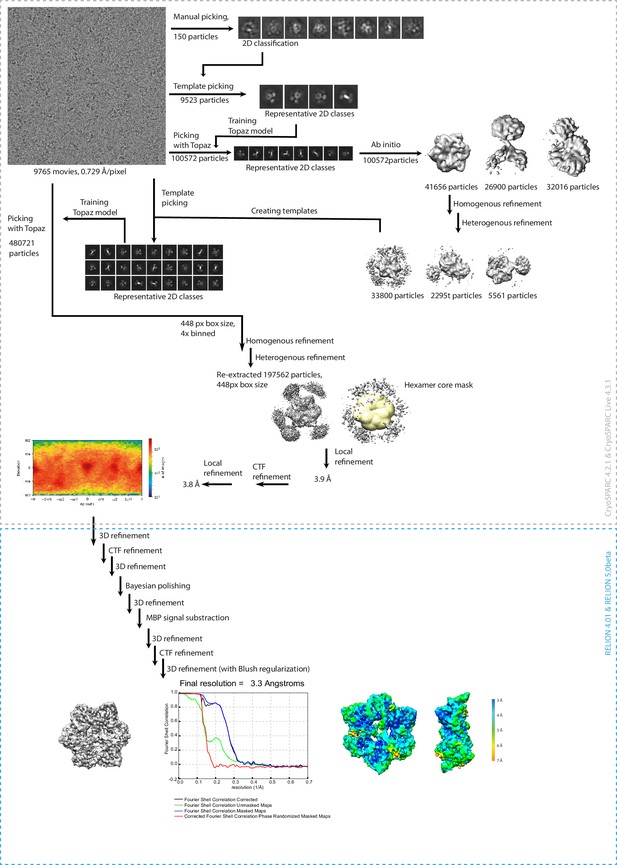
Flowchart illustrating cryo-EM data processing of MBP-fused CG67651-133.
Details are described in the Methods section.
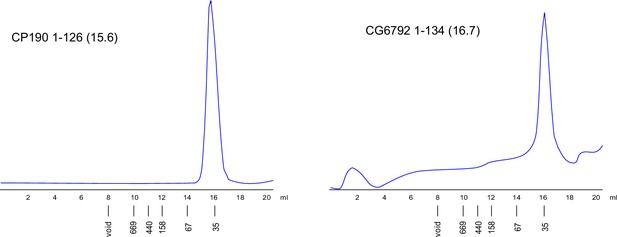
Superdex S200 size-exclusion chromatography of the CP190 and CG6792 broad-complex, tramtrack, and bric-a-brac (BTB) domains.
Positions of molecular weight markers are shown below. Molecular weights of monomers are shown in brackets.
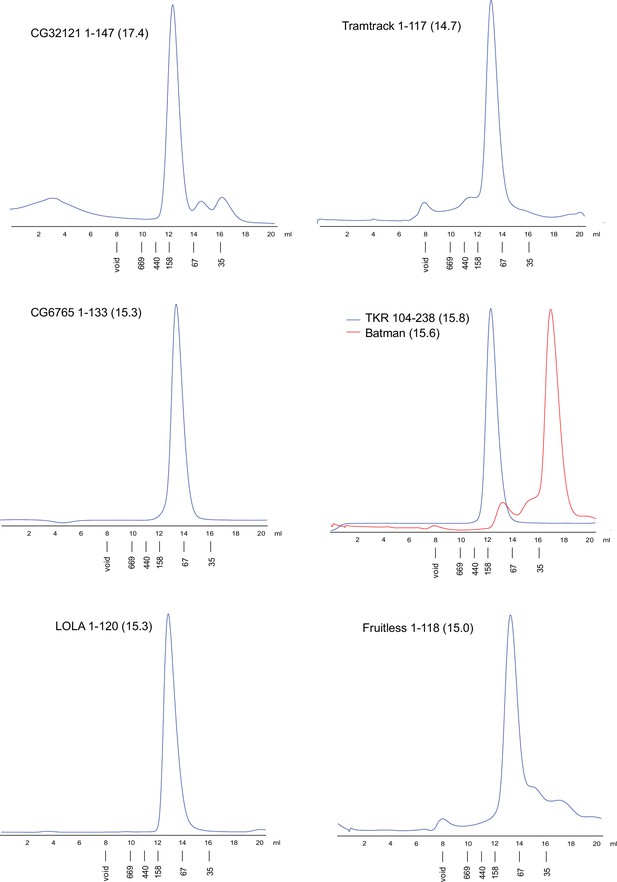
Superdex S200 size-exclusion chromatography of the Tramtrack group (TTK)-type broad-complex, tramtrack, and bric-a-brac (BTB) domains from Drosophila.
Positions of molecular weight markers (in kDa) are shown below. Molecular weights of monomers in kDa are shown in brackets.
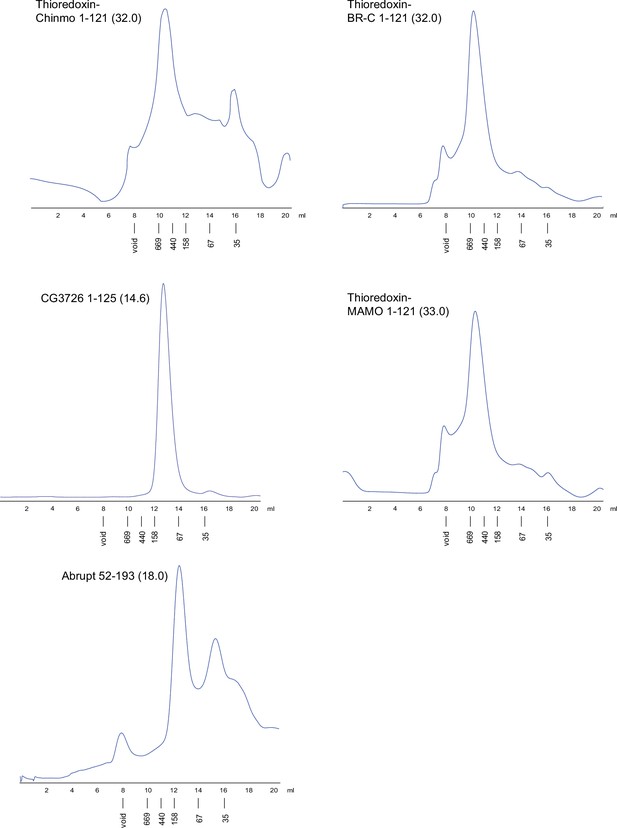
Superdex S200 size-exclusion chromatography of the Tramtrack group (TTK)-type broad-complex, tramtrack, and bric-a-brac (BTB) domains from Drosophila (continued).
Positions of molecular weight markers (in kDa) are shown below. Molecular weights of monomers in kDa are shown in brackets.
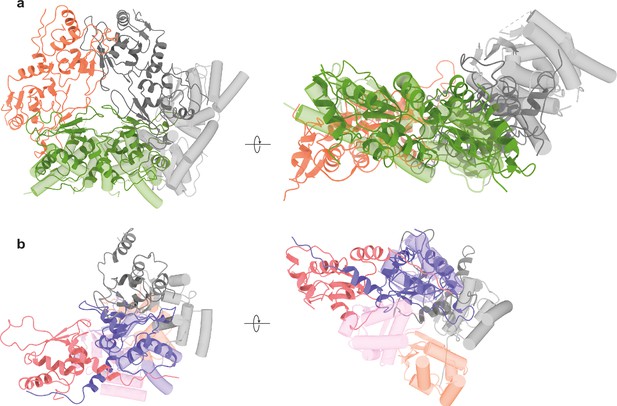
Overlays of multimeric assemblies of CG6765 hexamer with.
(a) ZBTB5 filamentous assembly (PDB ID: 9B9R Park et al., 2024) and with (b) T1-type tetrameric broad-complex, tramtrack, and bric-a-brac (BTB) from KV1.2 protein (PDB ID: 1QDV [Minor et al., 2000]). CG6765 hexamer is shown as ribbons, ZBTB5 /KV1.2 are shown as tube helices. Models are colored by dimer in (a) and by chain in (b). Only three subunits of CG6765 hexamer are shown in (b) for clarity.
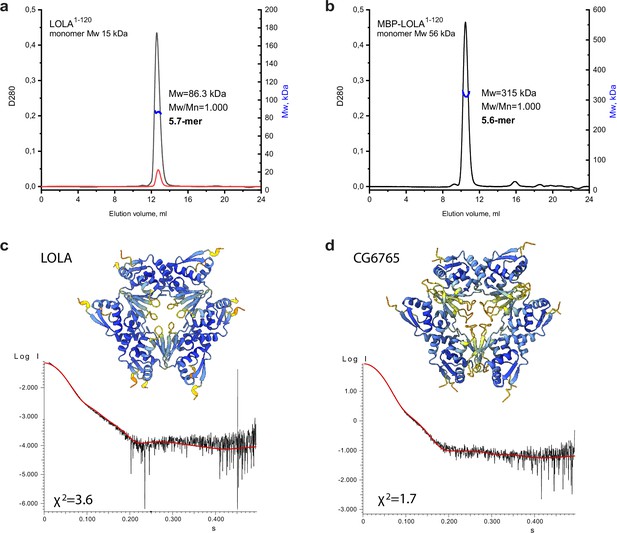
An integrative biology approach reveals the hexameric assembly of the Tramtrack group (TTK)-type broad-complex, tramtrack, and bric-a-brac (BTB) domains.
Size-exclusion chromatography with multi-angle light scattering (SEC-MALS) data for LOLA. (a) and MBP-LOLA (b) showing the chromatographic peaks with the Mw distributions across each peak. Average Mw values in kDa, polydispersity index (Mw/Mn), and the formally calculated oligomeric state are shown. D280 designates optical density at 280 nm (absorbance units). The second Y axes are Mw, and kDa. Note that the 10-fold dilution of LOLA (black and red curves in panel a) did not cause any shift of the chromatographic peak, indicating the stability of the observed oligomer. AlphaFold2-derived models of LOLA (c) and CG6765 (d) oligomers and fits of their theoretical scattering data to the experimental SAXS data. Models are colored according to AlphaFold pLDDT values.
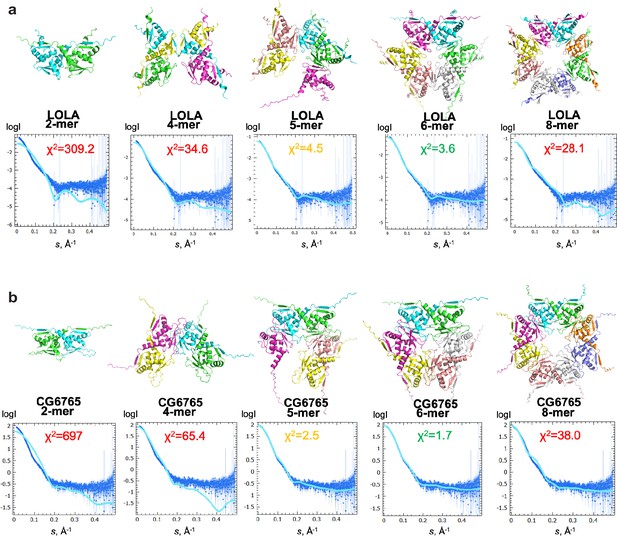
Studying the possibility of different stoichiometry of BTB domain assemblies using SAXS.
(a) Alphafold2 derived models of LOLA oligomers and their fits to the experimental SAXS data for the LOLA sample, shown along with the fit quality (chi2). (b) Alphafold2 derived models of CG6765 oligomers and their fits to the experimental SAXS data for the CG6765 sample, shown along with the fit quality (chi2). Note that the hexamers provided the most satisfactory fits. All attempts to build 7-mers returned disconnected models, which were not considered in analysis.
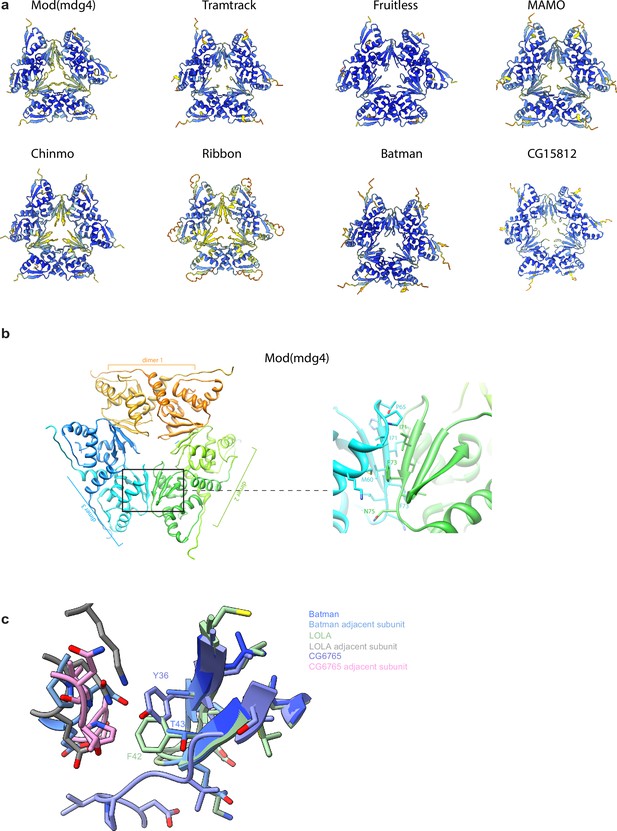
Molecular modeling of the Tramtrack group (TTK)-type broad-complex, tramtrack, and bric-a-brac (BTB) domains with AlphaFold.
(a) Models of hexameric assemblies of TTK-type BTB domains. Models are colored according to AlphaFold pLDDT values. (b) Molecular modeling of Mod(mdg4) BTB domain hexamer with a close-up view of dimer-dimer interaction interface. (c) Close-up view of dimer-dimer interaction interface of Alphafold models of LOLA, CG6765, and Batman hexamers.
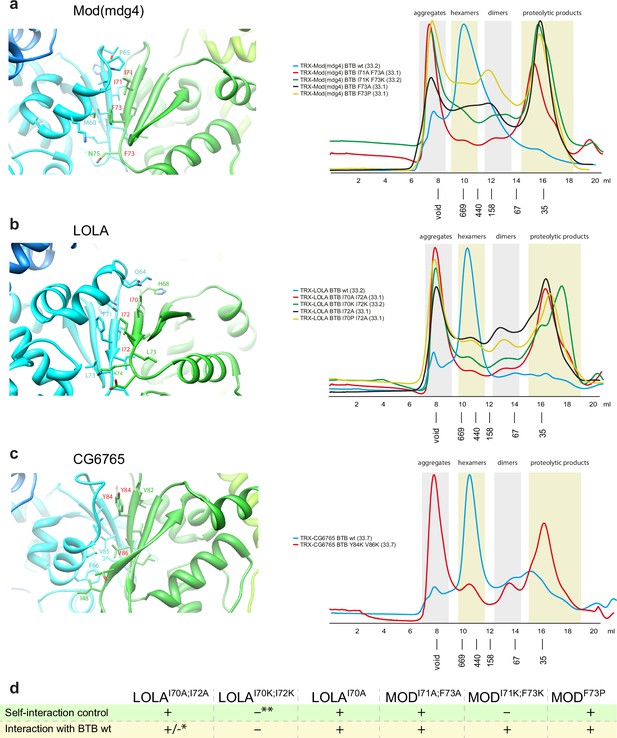
Testing the impact of single amino-acid substitutions in dimer-dimer interaction interface on the oligomerization status of the Tramtrack group (TTK)-type broad-complex, tramtrack, and bric-a-brac (BTB) domains.
Close-up views of AlphaFold-derived molecular models of dimer-dimer interaction interfaces and effect of single amino-acid substitutions on the oligomerization status of BTB domains of Mod(mdg4) (a), LOLA (b), and CG6765 (c) studied with Superdex S200 size-exclusion chromatography. Amino acids subjected to mutagenesis are shown in red. Positions of molecular weight markers are shown below. Molecular weights of monomers are shown in brackets. Further details are shown in Figure 4—figure supplements 4–6. (d) The self-interaction ability studied with a yeast two-hybrid assay. Growth assay plates are shown in Figure 4—figure supplement 7. asterisk indicates that the interaction was observed only in case when wild-type BTB was BD-fused. Double asterisk indicates that result is unreliable due to high self-activatory activity.
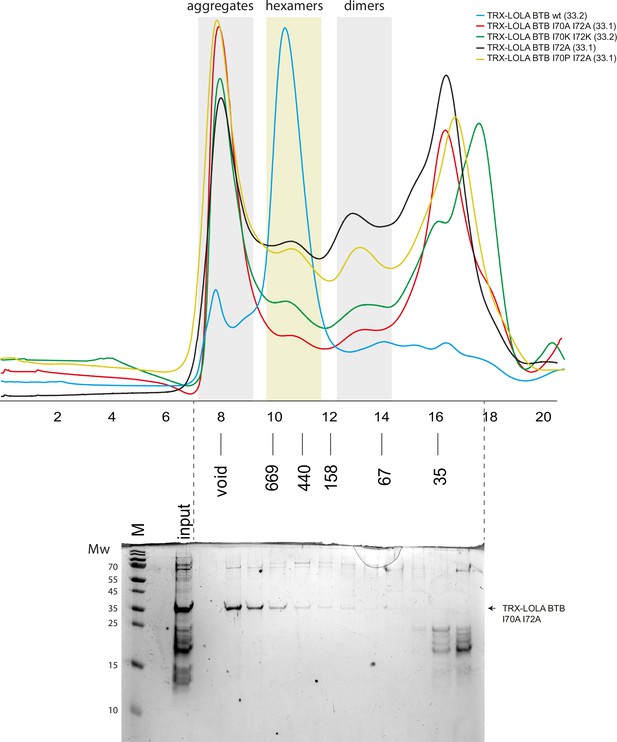
Superdex S200 size-exclusion chromatography of Thioredoxin-tagged LOLA broad-complex, tramtrack, and bric-a-brac (BTB) domains bearing mutations in dimer-dimer interaction interface.
Positions of molecular weight markers (in kDa) are shown below. Molecular weights of monomers in kDa are shown in brackets. The gel showing the content of size-exclusion chromatography (SEC) fractions peaks of alanine mutant is shown below, molecular weight markers are shown at the left. Uncropped gel images are available as separate files in Figure 4—figure supplement 4—source data 1.
-
Figure 4—figure supplement 4—source data 1
Uncropped gel images.
- https://cdn.elifesciences.org/articles/96832/elife-96832-fig4-figsupp4-data1-v2.zip
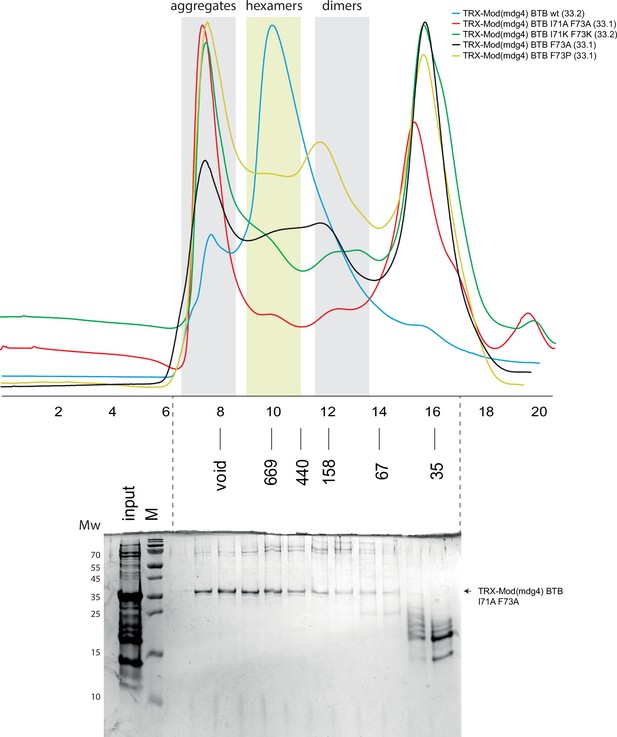
Superdex S200 size-exclusion chromatography of Thioredoxin-tagged Mod(mdg4) broad-complex, tramtrack, and bric-a-brac (BTB) domains bearing mutations in dimer-dimer interaction interface.
Positions of molecular weight markers (in kDa) are shown below. Molecular weights off monomers in kDa are shown in brackets. The gel showing the content of size-exclusion chromatography (SEC) fractions peaks of alanine mutant is shown below, molecular weight markers are shown at the left. Uncropped gel images are available as separate files in Figure 4—figure supplement 5—source data 1.
-
Figure 4—figure supplement 5—source data 1
Uncropped gel images.
- https://cdn.elifesciences.org/articles/96832/elife-96832-fig4-figsupp5-data1-v2.zip
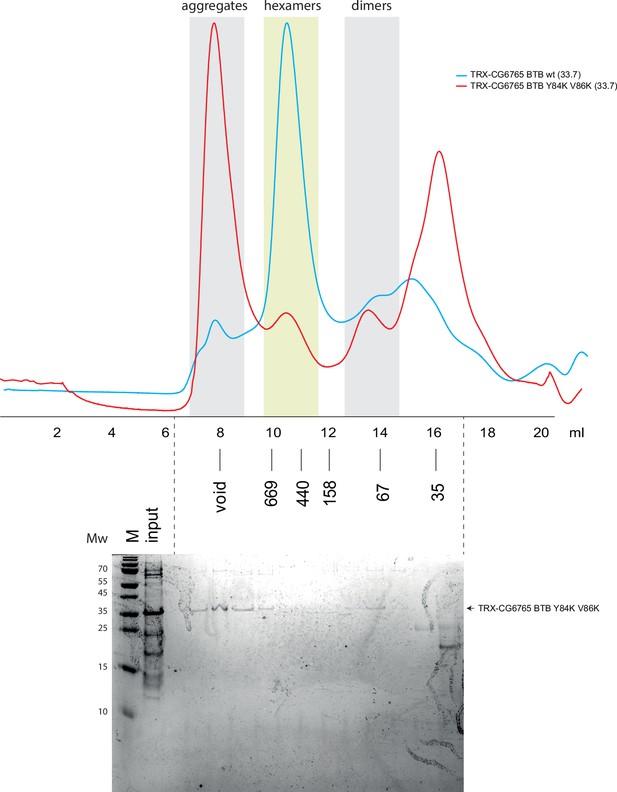
Superdex S200 size-exclusion chromatography of Thioredoxin-tagged CG6765 broad-complex, tramtrack, and bric-a-brac (BTB) domains bearing mutations in dimer-dimer interaction interface.
Positions of molecular weight markers (in kDa) are shown below. Molecular weights of monomers in kDa are shown in brackets. The gel showing the content of size-exclusion chromatography (SEC) fractions peaks of mutant protein is shown below, molecular weight markers are shown at the left. Uncropped gel images are available as separate files in Figure 4—figure supplement 6—source data 1.
-
Figure 4—figure supplement 6—source data 1
Uncropped gel images.
- https://cdn.elifesciences.org/articles/96832/elife-96832-fig4-figsupp6-data1-v2.zip
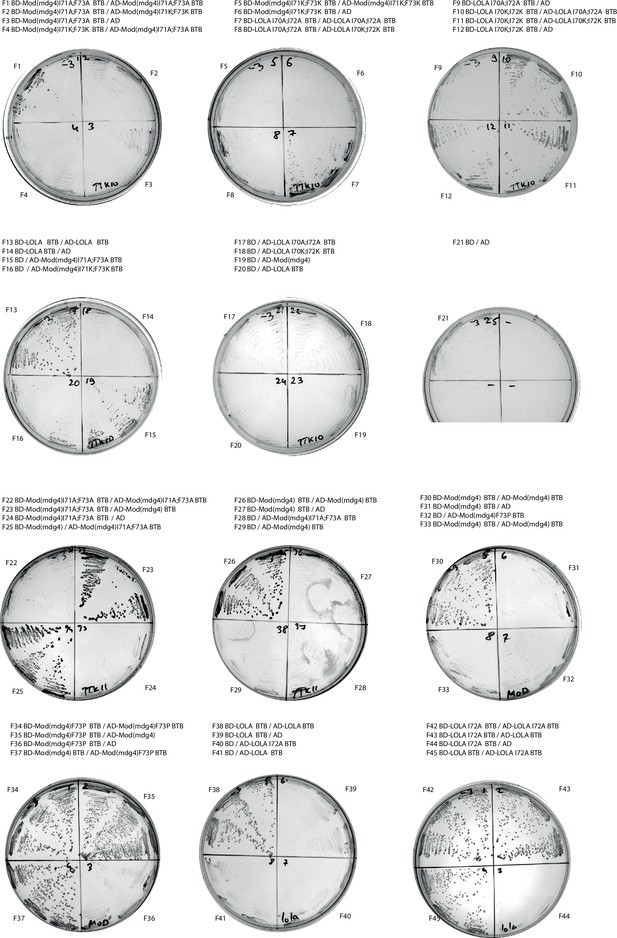
Results of yeast two-hybrid assays of the impact of point mutations at the dimer-dimer interaction interface.
Growth assay plates without histidine are shown (yeasts are unable to grow on this medium in the absence of interaction). AD stands for Activation Domain, BD – for DNA-Binding Domain of GAL4 protein.
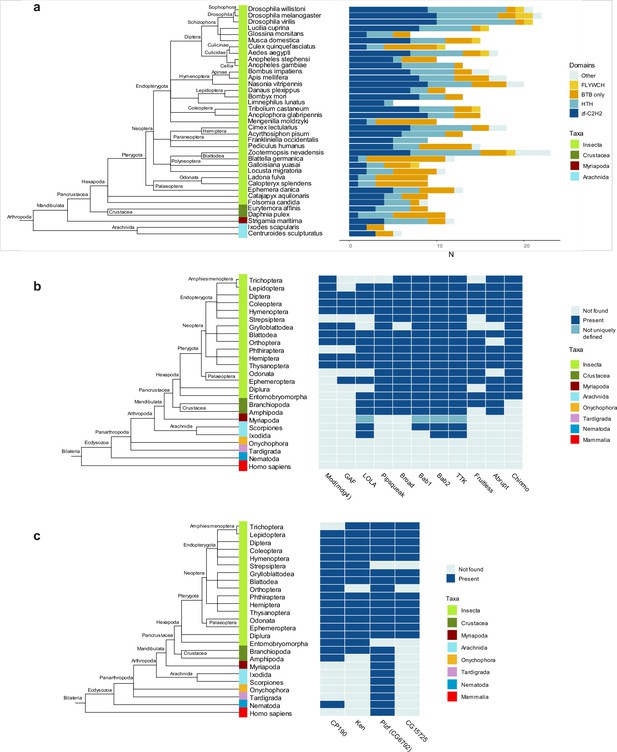
Domain architectures and orthologs of proteins with broad-complex, tramtrack, and bric-a-brac (BTB) domain of the Tramtrack group (TTK)-type in various Arthropoda lineages.
(a) Phylogenetic analysis of the distribution of the main DNA and protein interaction domain types in TTK-type BTB proteins in proteomes of representatives of Hexapoda, Crustacea, Myriapoda, and Arachnida. Each type of BTB protein domain architecture is shown as a bar segment. Total search results are shown in Supplementary file 7. The orthologs of several Drosophila BTB domains of TTK-type (b) and non-TTK-type (c) in proteomes of key taxa in major Arthropod and other Ecdysozoa phylogenetic groups. The phylogenetic relationships among taxa are according to the NCBI Taxonomy Database. Azure blue – the ortholog is absent in the taxa, dark blue – the ortholog is present in the taxa, dark cyan – orthologs are not uniquely defined.
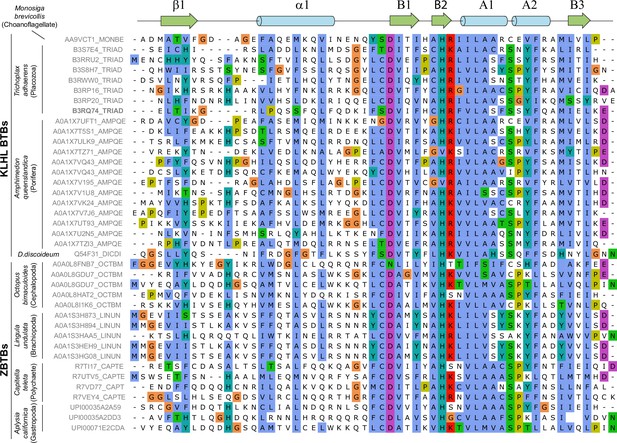
Multiple sequence alignment of β1-B3 sequences of broad-complex, tramtrack, and bric-a-brac (BTB) domains (KLHL family from basal Metazoans and ZBTB family from Protostomia clades beyond Arthropoda) bearing N-terminal beta-strand according to AlphaFold-multimer predictions. Amino acid residues are colored with ClustalX colors (legend is shown below).
BTB from Trichoplax B3RQ74 protein devoid of 3-box/BACK domain is shown in bold.
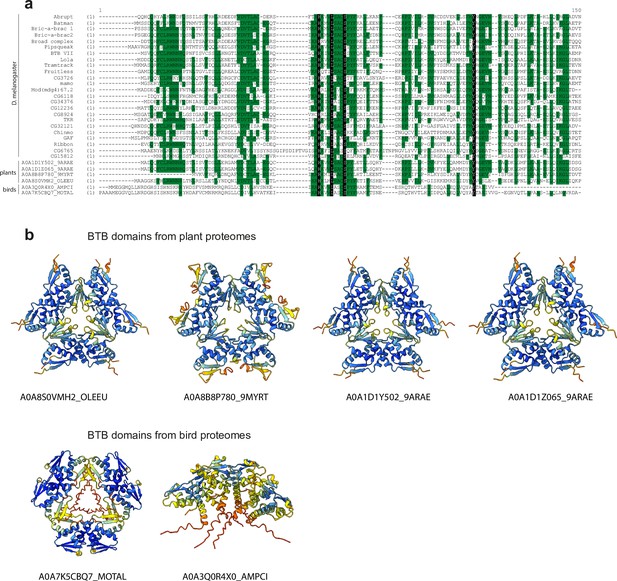
AlphaFold2 modelling of non-Arthropoda TTK-type BTB domains.
(a) Multiple sequence alignment of the Tramtrack group (TTK)-type broad-complex, tramtrack, and bric-a-brac (BTB) domains from Drosophila and top-score BTB domains found in InterPro using Hidden Markov Model (HMM) profile built on the sequences involved in the hexamer formation ranging from A2 to B3 structural elements. (b) AlphaFold-multimer predictions of top-score BTB domains from HMM search. Models are colored according to AlphaFold pLDDT values.
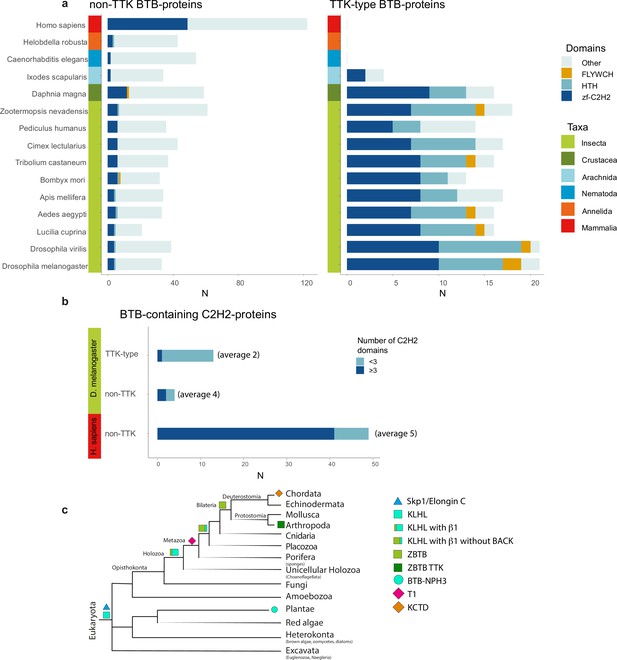
The Tramtrack group (TTK)-type broad-complex, tramtrack, and bric-a-brac (BTB) domains are specific to Arthropodan transcription factors.
(a) Distribution of major DNA interaction (C2H2, HTH, FLYWCH) and other domain types in non-TTK (left) and TTK-containing (right) BTB proteins in the proteomes of 16 Metazoan species. Each type of BTB-associated protein domain is denoted as a bar segment. Figure represents proteins with all BTB subtypes, domains of the ZBTB subtype are almost exclusively associated with one of the DNA-binding domains. Total search results are shown in Supplementary file 7 (TTK BTB domains) and Supplementary file 8 (non-TTK BTB domains).(b) Representation of non-TTK and TTK-containing BTB proteins with less than three (dark cyan) and three or more (dark blue) C2H2 domains in Drosophila melanogaster and Homo sapiens proteomes. (c) Origin of different subtypes of BTB domains over the course of evolution. (c) has been adapted from Figure 6C from Bonchuk et al., 2023.
Tables
SAXS-derived structural parameters for CG6765 and LOLA broad-complex, tramtrack, and bric-a-brac (BTB) domains.
Rg is the radius of gyration, Dmax – the maximum dimension of the particles, and Vp – is Porod volume – volume of the particles. The sample of CG67651-133 with the concentration of 1.5 mg/ml provided sufficiently high-quality scattering data, which were used for all fitting experiments, whereas data obtained for LOLA1-120 at concentrations 1.0 and 3.0 mg/ml were merged to obtain the necessary quality. Samples with higher concentrations exhibited signs of aggregation and were excluded from analysis.
| Polypeptide | Sample concentration, mg/ml | Rg, nm | Dmax, nm | Vp, nm3 | Estimated molecular weight, kDa | Molecular weight of the monomer, kDa |
|---|---|---|---|---|---|---|
| CG67651-133 | 1.5 | 3.7 | 12.7 | 166 | 90.5–116.5 | 15.3 |
| LOLA1-120 | merged data for 1.0 and 3.0 mg/ml | 4.1 | 14.5 | 189 | 108–136 | 15.2 |
| Reagent type (species) or resource | Designation | Source or reference | Identifiers | Additional information |
|---|---|---|---|---|
| Gene (Drosophila melanogaster) | CG6765 | GenBank | NM_139976.2 | |
| Gene (Drosophila melanogaster) | Lola | GenBank | NM_170623.6 | |
| Strain, strain background (Escherichia coli) | BL21(DE3) | Novagen | 69450 | |
| Recombinant DNA reagent | pMALX(A) (plasmid) | Moon et al., 2010 | ||
| Recombinant DNA reagent | pET32a(+) (plasmid) | Novagen | 69015 | |
| Recombinant DNA reagent | pGAD424 | TaKaRa bio | NCBI gi: 464015 | |
| Recombinant DNA reagent | pGBT9 | TaKaRa bio | NCBI gi: 470667 | |
| Software, algorithm | RELION 4.0 and 5.0beta | Scheres, 2012 Kimanius et al., 2023 | ||
| Software, algorithm | CryoSPARC v4.3.1 | Punjani et al., 2017 | ||
| Software, algorithm | PyEM | Asarnow et al., 2019 | ||
| Software, algorithm | UCSF Chimera | Pettersen et al., 2004 | ||
| Software, algorithm | Phenix.refine | Liebschner et al., 2019 | ||
| Software, algorithm | ISOLDE | Croll, 2018 | ||
| Software, algorithm | COOT | Emsley et al., 2010 | ||
| Software, algorithm | Topaz | Bepler et al., 2019 | ||
| Software, algorithm | ChimeraX v.1.6 | Pettersen et al., 2021 | ||
| Software, algorithm | ATSAS package | Franke et al., 2017 | ||
| Software, algorithm | AlphaFold2 | Evans et al., 2022 | ||
| Software, algorithm | AlphaPulldown | Yu et al., 2023 | ||
| Software, algorithm | HMMSEARCH | Potter et al., 2018 | ||
| Software, algorithm | MEME | Bailey et al., 2009 | ||
| Software, algorithm | MUSCLE | Edgar, 2004 | ||
| Software, algorithm | biomaRt | Durinck et al., 2009 | ||
| Software, algorithm | OrthoDB | Kriventseva et al., 2019 | ||
| Software, algorithm | MEGA-X | Kumar et al., 2018 | ||
| Software, algorithm | taxize | Chamberlain and Szöcs, 2013 | ||
| Software, algorithm | CDD/SPARCLE NCBI | Lu et al., 2020 |
Additional files
-
MDAR checklist
- https://cdn.elifesciences.org/articles/96832/elife-96832-mdarchecklist1-v2.docx
-
Supplementary file 1
Cryo-EM data collection and processing statistics.
- https://cdn.elifesciences.org/articles/96832/elife-96832-supp1-v2.docx
-
Supplementary file 2
Summary of the ability of GAF, Mod(mdg4), Chinmo, CG8924 and LOLA BTB domains to interact with other TTK-type BTB domains in yeast two-hybrid assay.
- https://cdn.elifesciences.org/articles/96832/elife-96832-supp2-v2.docx
-
Supplementary file 3
Pairwise level of similarity between GAF and Mod(mdg4) BTB domains with other TTK-type domains.
- https://cdn.elifesciences.org/articles/96832/elife-96832-supp3-v2.docx
-
Supplementary file 4
Testing of the interactions between the non-TTK BTB domains of C2H2 proteins.
- https://cdn.elifesciences.org/articles/96832/elife-96832-supp4-v2.docx
-
Supplementary file 5
The ability of non-ttk BTB domains of CP190, CG6792, CG15725 and Ken proteins to interact with other TTK-type BTB domains in yeast two-hybrid assay.
- https://cdn.elifesciences.org/articles/96832/elife-96832-supp5-v2.docx
-
Supplementary file 6
AlphaFold2-Multimer – modeled heteromeric interactions of TTK-group BTB domains with interaction confirmed in Y2H assay.
- https://cdn.elifesciences.org/articles/96832/elife-96832-supp6-v2.docx
-
Supplementary file 7
Total domain search results for proteins with TTK-type BTB domains.
- https://cdn.elifesciences.org/articles/96832/elife-96832-supp7-v2.xlsx
-
Supplementary file 8
Total domain search results for proteins with non-TTK-type BTB domains.
- https://cdn.elifesciences.org/articles/96832/elife-96832-supp8-v2.xlsx
-
Supplementary file 9
Total search results for HMM-profile built on consensus sequence of dimer-dimer interaction interface.
- https://cdn.elifesciences.org/articles/96832/elife-96832-supp9-v2.xlsx
-
Supplementary file 10
Oligonucleotides used for cloning.
- https://cdn.elifesciences.org/articles/96832/elife-96832-supp10-v2.docx





