Base editing of Ptbp1 in neurons alleviates symptoms in a mouse model of Parkinson’s disease
Figures
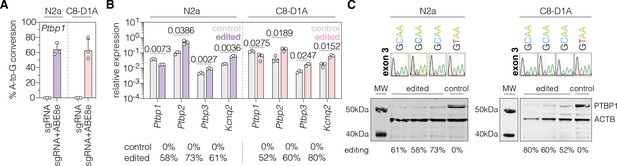
Polypyrimidine tract binding protein 1 (PTBP1) downregulation by adenine base editing with sgRNA-ex3 in neuronal and astroglial cell lines.
(A) Editing rates at the Ptbp1 splice donor of exon 3 in N2a and C8-D1A cell lines. Editing efficiencies were determined by Sanger sequencing and EditR (Kluesner et al., 2018). Control samples were transfected with sgRNA (gray). (B) Transcript levels of Ptbp1 and Ptbp1-repressed exons upon adenine base editing in N2a and C8-D1A cells. Transcripts were normalized to Gapdh. (C) PTBP1 levels in control (1 independent experiment) and edited N2a or C8-D1A cells (3 independent experiments). ACTB protein levels are shown as a loading control. Corresponding sequencing chromatograms for sgRNA-ex3 are shown above each sample. Corresponding editing rates are shown below the plots in (B) and (C). Normal distribution of the data was analyzed using the Shapiro-Wilk test. Data are represented as means ± s.d. of three independent experiments (A,B) and were analyzed using an unpaired two-tailed Student’s t-test with Welch’s correction (B). Each datapoint represents one independent experiment. Exact P-values are indicated in the respective plots. Ptbp1, Polypyrimidine tract binding protein 1; Ptbp2, Polypyrimidine tract binding protein 2; Ptbp3, Polypyrimidine tract binding protein 3; Kcnq2, potassium voltage-gates channel subfamily Q member 2; MW, molecular weight marker; kDa, kilodalton.
-
Figure 1—source data 1
Original membranes corresponding to Figure 1 (panel C).
Size of molecular weight (MW) markers are indicated. Bands for PTBP1 (57 kDa) and beta-actin (45 kDa) are indicated for sgRNA-ex3 in N2a and C8-D1A cells.
- https://cdn.elifesciences.org/articles/97180/elife-97180-fig1-data1-v1.zip
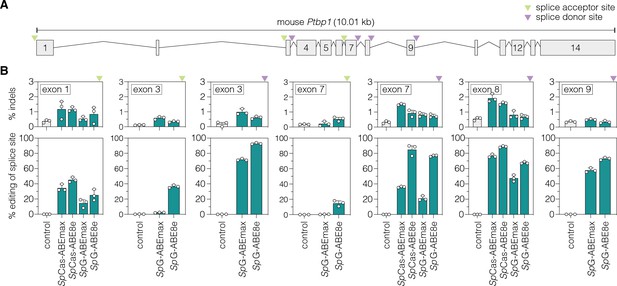
Adenine base editing of Ptbp1 splice sites in murine Hepa cells.
(A) Schematic overview of exons and introns of the targeted murine Ptbp1 locus. sgRNAs target either the conserved GT motif of canonical splice donor sites at the beginning of an intron (purple arrowhead) or the conserved AG motif of canonical splice acceptor sites at the end of an intron (green arrowhead). (B) Editing of adenines and indel rates at Ptbp1 splice donor (purple arrowhead) or acceptor sites (green arrowhead) for SpCas- or SpG-ABE variants (teal). The targeted exons are indicated in the respective plot. Control samples were transfected with sgRNA (gray). Data are displayed as means ± s.d. of three independent experiments. Each datapoint represents one independent experiment. Ptbp1, polypyrimidine tract binding protein 1; kb, kilobases; SpCas, Streptococcus pyogenes Cas9 recognizing NGG PAMs; SpG, SpCas variant recognizing NGN PAMs.
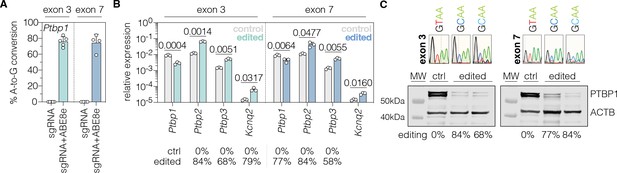
In vitro validation of PTBP1 repression by adenine base editing with sgRNA-ex3 or sgRNA-ex7 in Hepa cells.
(A) Editing rates at Ptbp1 splice donor sites of exon 3 and exon 7. Editing efficiencies were determined by Sanger sequencing and EditR (Kluesner et al., 2018). Control samples were transfected with sgRNA (gray). (B) Transcript levels of Ptbp1 and Ptbp1-repressed exons upon adenine base editing in Hepa cells. Transcripts were normalized to Gapdh. (C) PTBP1 levels in control (1 independent experiment) and edited Hepa cells (two independent experiments). ACTB protein levels are shown as a loading control. Corresponding sequencing chromatograms for sgRNA-ex3 and sgRNA-ex7 are shown above each sample. Corresponding editing rates are shown below the plots in (B) and (C). Normal distribution of the data was analyzed using the Shapiro-Wilk test. Data are represented as means ± s.d. of at least three independent experiments (A, B) and were analyzed using an unpaired two-tailed Student’s t-test with Welch’s correction (B). Each datapoint represents one independent experiment. Exact p-values are indicated in the respective plots. Ptbp1, Polypyrimidine tract binding protein 1; Ptbp2, Polypyrimidine tract binding protein 2; Ptbp3, Polypyrimidine tract binding protein 3; Kcnq2, potassium voltage-gates channel subfamily Q member 2; ctrl, control; MW, molecular weight marker.
-
Figure 1—figure supplement 2—source data 1
Original membranes corresponding to Figure 1—figure supplement 2 (panel C).
Size of molecular weight (MW) markers are indicated. Bands for polypyrimidine tract binding protein 1 (PTBP1) (57 kDa) and beta-actin (45 kDa) are indicated for sgRNA-ex3 and sgRNA-ex7 in Hepa1-6 cells.
- https://cdn.elifesciences.org/articles/97180/elife-97180-fig1-figsupp2-data1-v1.zip
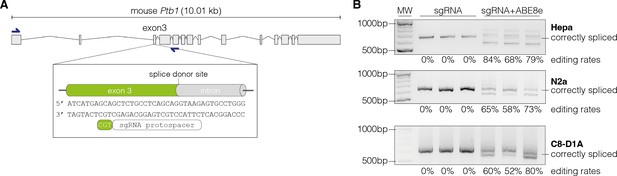
Adenine base editing generates alternative polypyrimidine tract binding protein 1 (Ptbp1) splice sites in cell lines.
(A) Schematic representation of the splice donor at the exon-intron junction of Ptbp1 exon 3. (B) Editing of the canonical splice donor at exon 3 generates alternative Ptbp1 splice sites in Hepa, N2a, and C8-D1A cells. The correctly spliced isoform is labeled. The corresponding editing rates are shown below each plot. Data from three independent experiments are shown (B). Size of molecular weight (MW) markers in base pairs (bp) are indicated in (B).
-
Figure 1—figure supplement 3—source data 1
Original gel images corresponding to Figure 1—figure supplement 3 (panel B).
Size of molecular weight (MW) markers are indicated. Bands for correctly spliced isoforms are indicated.
- https://cdn.elifesciences.org/articles/97180/elife-97180-fig1-figsupp3-data1-v1.zip
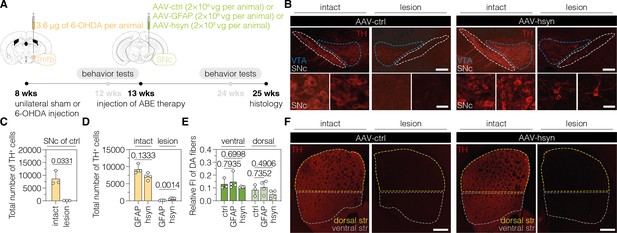
Downregulation of polypyrimidine tract binding protein 1 (PTBP1) in neurons of the SNc generates TH+ cells.
(A) Schematic representation of the experimental timeline and setup. (B) Representative images of midbrain sections showing the intact (left) or lesioned (right) SNc in animals treated with adeno-associated virus (AAV)-ctrl (left) or AAV-hsyn (right). Treatment groups and hemispheres are indicated on top. (C, D) Quantification of TH+ cells in the intact or lesioned SNc in animals treated with AAV-ctrl (C), AAV-GFAP (D), or AAV-hsyn (D) in the lesioned hemisphere. (E,F) Quantifications (E) of DA fibers in the striatum, assessed as relative fluorescence intensity (FI) of TH compared to the intact striatum of the same section, and representative images of brain sections (F) showing the intact or denervated striatum (str) in animals treated with AAV-ctrl (left) or AAV-hsyn (right). The FI of the TH staining detected in the corpus callosum of each hemisphere was used for background correction of FI detected in the striatum of the same hemisphere. Control animals were treated with AAV-PHP.eB particles, expressing the ABE8e variant under the ubiquitous Cbh promoter. Tissue areas used for quantifications are marked by colored dashed lines in (B) and (E). Normal distribution of the data was analyzed using the Shapiro-Wilk test. Data are represented as means ± s.d. of 3–8 animals per group and were analyzed using an unpaired two-tailed Student’s t-test with Welch’s correction (C, D) or a one-way ANOVA with Dunnett’s multiple comparisons test (E). Each datapoint represents one animal. Exact p-values are indicated in the respective plots. Scale bars, 20 μm (B, bottom) and 1000 μm (B, top; F). ctrl, AAV-ctrl-ABE treatment; GFAP, AAV-GFAP-ABE treatment; hsyn, AAV-hsyn-ABE treatment; ABE, adenine base editor; vg, vector genomes; SNc, substantia nigra pars compacta; VTA, ventral tegmental area; TH, tyrosine hydroxylase; str, striatum; FI, fluorescence intensity; DA, dopaminergic.
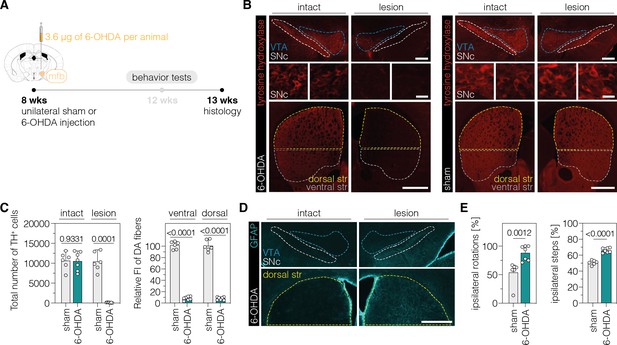
Validation of the unilateral 6-OHDA lesion in C57BL/6 J mice.
(A) Schematic representation of the experimental timeline and setup. (B) Representative fluorescence images showing the unilateral loss of TH+ cells in the SNc (top and middle) and unilateral depletion of DA fibers in the striatum (bottom) in 6-OHDA-lesioned mice (left). Sham-injected animals (right) are shown for comparison. The FI of the TH staining detected in the corpus callosum of each hemisphere was used for background correction of FI detected in the striatum of the same hemisphere. Treatment groups are indicated on the left. (C) Quantifications of TH+ DANs in the SNc (left) and DA fibers in the ventral and dorsal striatum (right). Tissue areas used for quantifications are marked by colored dashed lines in (B). (D) Unilateral activation of astrocytes in the SNc (top) and dorsal striatum (bottom) in 6-OHDA-lesioned mice. (E) Spontaneous behaviors in sham- and 6-OHDA-lesioned mice. Normal distribution of the data was analyzed using the Shapiro-Wilk test. Data are displayed as means ± s.d. of 6–7 mice per group and were analyzed using an unpaired two-tailed Student’s t-test with Welch’s correction (C, ipsilateral steps in E) or a two-tailed Mann Whitney test (ipsilateral rotations in E). Each datapoint represents one animal. Exact P-values are indicated in the respective plots. Scale bars, 20 μm (B, bottom) and 1000 μm (B, top; D). FI, fluorescence intensity; SNc, substantia nigra pars compacta; str, striatum; VTA, ventral tegmental area; GFAP, glial fibrillary acidic protein; TH, tyrosine hydroxylase; DA, dopaminergic.
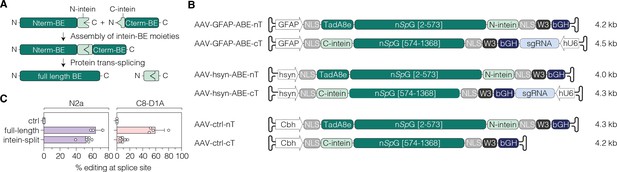
Adeno-associated virus (AAV) vector designs for neuronal or astroglial expression of intein-split ABE8e.
(A) Depiction of Npu intein-split BE moieties (Nterm-BE and Cterm-BE), forming the full length BE after protein trans-splicing. (B) Schematic representation of AAV vector designs with the short GFAP (Lee et al., 2008), hsyn (Kügler et al., 2003), or Cbh promoter (Gray et al., 2011) and their corresponding lengths in kilobase pairs (including ITRs). Constructs are not depicted to scale. (C) Editing efficiencies of full-length (CMV promoter) or intein-split ABE expression vectors in N2a (hsyn promoter) and C8-D1A cells (GFAP promoter). Control samples were treated with the sgRNA only. Data from 3–6 independent experiments are displayed as means ±s.d. (C). BE, base editor; ABE, adenine base editor; nT/Nterm, N-terminal AAV construct; cT/Cterm, C-terminal AAV construct; GFAP, glial fibrillary acidic protein promoter; hsyn, human synapsin 1 promoter; Cbh, truncated chimeric CMV/chicken b-actin hybrid promoter; NLS, nuclear localization signal; TadA8e, adenosine deaminase; nSpG, SpG nickase; W3, woodchuck hepatitis virus post-transcriptional regulatory element; bGH, bovine growth hormone polyadenylation signal; hU6, human U6 promoter; kb, kilobase pairs.
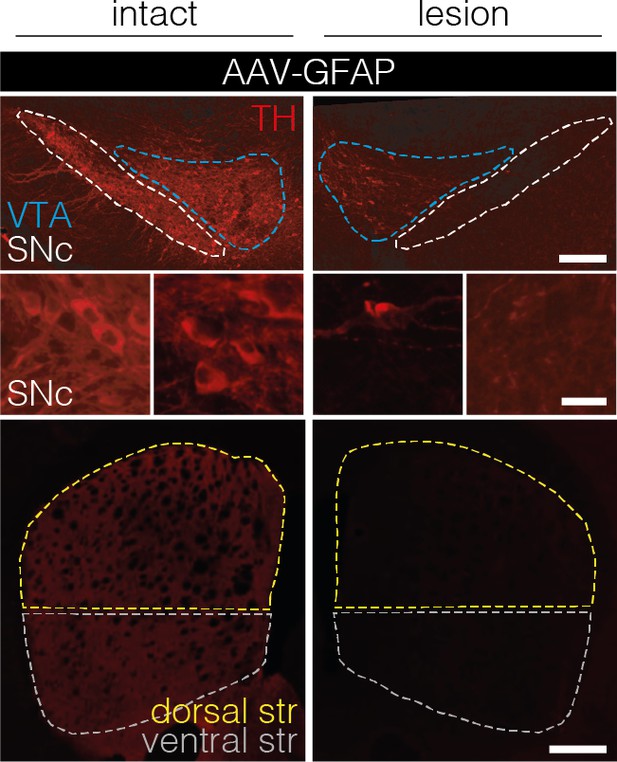
Polypyrimidine tract binding protein 1 (PTBP1) downregulation in astrocytes of the SNc fails to generate TH+ cells.
Representative images of brain sections showing the intact (left) or lesioned (right) SNc (top and middle) or striatum (bottom) in animals after astroglial PTBP1 downregulation (n=4 mice). The fluorescence intensity (FI) of the TH staining detected in the corpus callosum of each hemisphere was used for background correction of FI detected in the striatum of the same hemisphere. Tissue areas used for quantifications are marked by colored dashed lines. Scale bars, 20 μm (middle) and 1000 μm (top and bottom). AAV-GFAP, AAV-GFAP-ABE treatment; GFAP, glial fibrillary acidic protein; SNc, substantia nigra pars compacta; VTA, ventral tegmental area; TH, tyrosine hydroxylase.
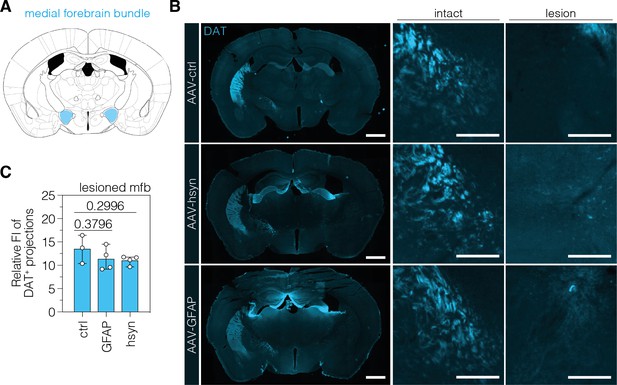
Tyrosine hydroxylase (TH)-expressing cells in the substantia nigra pars compacta (SNc) do not form neuronal projections through the mfb.
(A, B) Schematic depiction of the mfb (A) on the mouse brain atlas (Paxinos and Franklin, 2001) and representative images of DA projections in the mfb of the intact and lesioned hemisphere in treated Parkinson’s disease (PD) mice. Treatment groups (left) and hemispheres (top) are indicated. (C) Quantifications of DA projections in the mfb, assessed as relative fluorescence intensity (FI) of the dopaminergic marker DAT (dopamine transporter) compared to the intact mfb of the same section, in animals treated with AAV-ctrl, AAV-GFAP, or AAV-hsyn. The FI of the DAT staining detected in the thalamus of each hemisphere was used for background correction of FI detected in the mfb of the same hemisphere. Control animals were treated with AAV-PHP.eB particles, expressing the ABE8e variant under the ubiquitous Cbh promoter. Normal distribution of the data was analyzed using the Shapiro-Wilk test. Data are represented as means ± s.d. of 3–4 animals per group and were analyzed using a one-way ANOVA with Dunnett’s multiple comparisons test. Each datapoint represents one animal. Exact p-values are indicated in the respective plots. Scale bars, 1000 μm (left) and 50 μm (middle and right). ctrl, AAV-ctrl-ABE treatment; GFAP, AAV-GFAP-ABE treatment; hsyn, AAV-hsyn-ABE treatment; mfb, medial forebrain bundle; FI, fluorescence intensity; DA, dopaminergic; DAT, dopamine transporter.

In vivo validation of polypyrimidine tract binding protein 1 (PTBP1) downregulation by adenine base editing in astrocytes and neurons of the SNc.
(A) Quantifications of in vivo base editing of adenines and indel rates within the editing window at the targeted Ptbp1 splice donor. (B) Transcript levels of Ptbp1 upon adenine base editing in the lesioned SNc of AAV-ctrl-, AAV-GFAP-, and AAV-hsyn-treated Parkinson’s disease (PD) mice. Transcripts were normalized to Gapdh. (C) PTBP1 levels in the lesioned SNc of treated 6-OHDA-lesioned PD mice (n=3 mice per group). ACTB protein levels are shown as a loading control. PTBP1 abundance was compared to ACTB and normalized to the PTBP1/ACTB ratio of AAV-ctrl-treated animals. Control animals were treated with AAV-PHP.eB particles, expressing the ABE8e variant under the ubiquitous Cbh promoter. Normal distribution of the data was analyzed using the Shapiro-Wilk test. Data are represented as means ± s.d. of 3–7 animals per group and were analyzed using a one-way ANOVA with Dunnett’s multiple comparisons test (C). Each datapoint represents one animal. Exact p-values are indicated in the respective plots. ctrl, AAV-ctrl-ABE treatment; GFAP, AAV-GFAP-ABE treatment; hsyn, AAV-hsyn-ABE treatment; SNc, substantia nigra pars compacta; Ptbp1/PTBP1, Polypyrimidine tract binding protein 1; ACTB, beta actin; MW, molecular weight marker.
-
Figure 2—figure supplement 5—source data 1
Original membranes corresponding to Figure 2—figure supplement 5 (panel C).
Size of molecular weight (MW) markers are indicated. Bands for polypyrimidine tract binding protein 1 (PTBP1) (57 kDa) and beta-actin (45 kDa) are indicated in the substantia nigra pars compacta (SNc) of Parkinson’s disease (PD) mice.
- https://cdn.elifesciences.org/articles/97180/elife-97180-fig2-figsupp5-data1-v1.zip
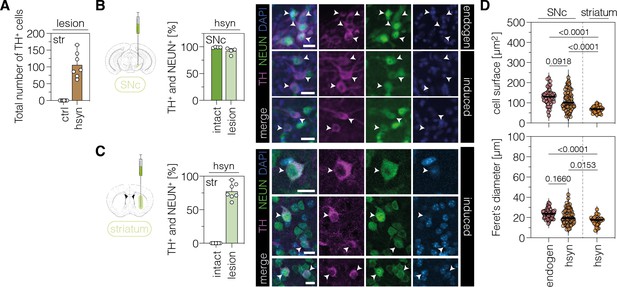
Characterization of TH-expressing cells in the SNc or striatum.
(A) Quantification of TH+ cells in the lesioned striatum of animals treated with AAV-ctrl or AAV-hsyn. Control animals were treated with AAV-PHP.eB particles, expressing the ABE8e-SpG variant under the ubiquitous Cbh promoter. (B,C) Quantifications (left) and representative images (right) of TH/NeuN double-positive cell bodies in the intact (dark green, labeled as ‘endogen’ in the images) or lesioned (light green, labeled as ‘induced’ in the images) SNc (B) or striatum (C) of AAV-hsyn-treated animals. (D) Corresponding quantifications of cell surface area and Feret’s diameter (longest distance between the cell boundaries) of TH/NeuN double-positive cell bodies in the intact (labeled as ‘endogen’) or lesioned SNc or striatum (labeled as ‘hsyn’). Normal distribution of the data was analyzed using the Shapiro-Wilk test. Data are displayed as means ± s.d. of 4–7 mice per group (A–C) or 32–68 TH/NeuN double-positive cells per group (D; n=4 mice) and were analyzed using a one-way ANOVA with Tukey’s multiple comparisons test (D). Each datapoint represents one animal (A–C) or a TH/NeuN double-positive cell (D). Exact P-values are indicated in the each plot above the respective group (D). Scale bars, 20 μm. ctrl, AAV-ctrl-ABE treatment; hsyn, AAV-hsyn-ABE treatment; SNc, substantia nigra pars compacta; str, striatum; TH, tyrosine hydroxylase; NEUN, hexaribonucleotide binding protein-3; DAPI, 4’,6-diamidino-2-phenylindole; endogen, endogenous.
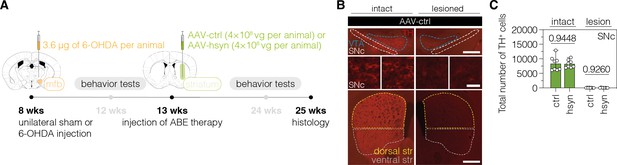
Validation of the unilateral lesion in 6-OHDA mice after neuronal polypyrimidine tract binding protein 1 (PTBP1) downregulation in the striatum.
(A) Schematic representation of the experimental timeline and setup. (B) Representative fluorescence images showing the unilateral loss of TH+ cells in the SNc (top and middle) and unilateral depletion of DA fibers in the striatum (bottom) in 6-OHDA-lesioned mice treated with AAV-ctrl. Scale bars, 1000 μm (top and bottom) and 20 μm (middle). (C) Quantifications of TH+ cells in the intact (dark green) and lesioned (light green) SNc after administration of the AAV-hsyn treatment to the striatum. Control animals were treated with AAV-PHP.eB particles, expressing the ABE8e-SpG variant under the Cbh promoter. Tissue areas used for quantifications are marked by colored dashed lines in (B). Normal distribution of the data was analyzed using the Shapiro-Wilk test. Data are displayed as means ± s.d. of 8 mice per group and were analyzed using an unpaired two-tailed Student’s t-test with Welch’s correction (C, ‘intact’) or a two-tailed Mann-Whitney test (C, ‘lesion’). Each datapoint represents one animal. Exact p-values are indicated in the respective plots. ABE, adenine base editor; ctrl, AAV-ctrl-ABE treatment; hsyn, AAV-hsyn-ABE treatment; SNc, substantia nigra pars compacta; VTA, ventral tegmental area; TH, tyrosine hydroxylase.
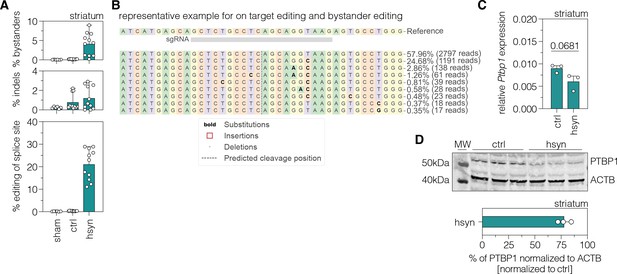
In vivo validation of polypyrimidine tract binding protein 1 (PTBP1) downregulation by adenine base editing in neurons of the striatum.
(A) Quantifications of in vivo base editing of adenines and indel rates within the editing window at the targeted Ptbp1 splice donor (n=6–11 mice per group). (B) Representative CRISPResso output file showing the frequency and positioning of edited adenines in the lesioned striatum. (C) Transcript levels of Ptbp1 upon adenine base editing in the lesioned striatum. Transcripts were normalized to Gapdh (n=3 mice per group). (D) PTBP1 levels in the lesioned striatum of treated 6-OHDA-lesioned PD mice (n=3 mice per group). ACTB protein levels are shown as a loading control. PTBP1 abundance was compared to ACTB and normalized to the PTBP1/ACTB ratio of AAV-ctrl-treated animals. Control animals were treated with AAV-PHP.eB particles, expressing the ABE8e variant under the ubiquitous Cbh promoter. Normal distribution of the data was analyzed using the Shapiro-Wilk test. Data are represented as means ± s.d. of 3–11 animals per group and were analyzed using an unpaired two-tailed Student’s t-test with Welch’s correction (B). Each datapoint represents one animal. Exact p-values are indicated in the respective plots. ctrl, AAV-ctrl-ABE treatment; hsyn, AAV-hsyn-ABE treatment; str, striatum; Ptbp1/PTBP1, Polypyrimidine tract binding protein 1; ACTB, beta actin; MW, molecular weight marker.
-
Figure 3—figure supplement 2—source data 1
Original membranes corresponding to Figure 3—figure supplement 2 (panel D).
Size of molecular weight (MW) markers are indicated. Bands for polypyrimidine tract binding protein 1 (PTBP1) (57 kDa) and beta-actin (45 kDa) are indicated in the striatum of Parkinson’s disease (PD) mice.
- https://cdn.elifesciences.org/articles/97180/elife-97180-fig3-figsupp2-data1-v1.zip
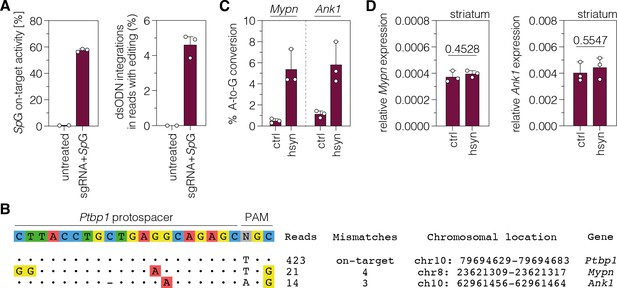
Identification of sgRNA-dependent off-target sites using GUIDE-seq.
(A) Frequency of indels of sgRNA-ex3 and the SpG nuclease (left) and dsODN integrations within the edited reads (right) in N2a cells. (B) Experimentally identified off-target sites for the sgRNA-ex3 protospacer targeting the polypyrimidine tract binding protein 1 (Ptbp1) locus. The number of reads for the on-target (top) and off-target sites (middle and bottom) as well as the number of mismatches, chromosomal location, and gene annotation are indicated on the right. (C, D) Deep sequencing results of the off-target sites (C) and transcript levels (D) of Mypn and Ank1 upon adenine base editing in the lesioned striatum. Transcripts were normalized to Gapdh (n=3 mice per group). Data are represented as means ± s.d. of three animals per group and were analyzed using an unpaired two-tailed Student’s t-test with Welch’s correction (D).
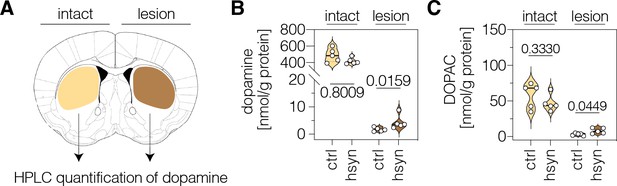
Neuronal polypyrimidine tract binding protein 1 (PTBP1) repression in the striatum increases striatal dopamine in 6-OHDA-lesioned hemispheres.
(A) Schematic representation of striatal regions used for neurotransmitter quantifications. (B, C) HPLC quantifications of dopamine (B) and its metabolite DOPAC (C) in the intact and lesioned striatum of animals treated with AAV-ctrl or AAV-hsyn. Control animals were treated with AAV-PHP.eB particles, expressing the ABE8e-SpG variant under the Cbh promoter. Neurotransmitter concentrations in nmol were normalized to total protein amounts in g. Normal distribution of the data was analyzed using the Shapiro-Wilk test. Data are displayed as means ± s.d. of five mice per group and were analyzed using an unpaired two-tailed Student’s t-test (‘intact’ in B; C) or a two-tailed Mann-Whitney test (‘lesion’ in B). Each datapoint represents one animal. Exact p-values are indicated in the respective plots. ctrl, AAV-ctrl-ABE treatment; hsyn, AAV-hsyn-ABE treatment; DOPAC, 3,4-dihydroxyphenylacetic acid; nmol, nanomol; g, gram.
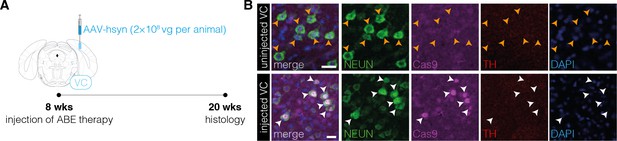
Absence of TH+ cell bodies in the visual cortex after neuronal polypyrimidine tract binding protein 1 (PTBP1) downregulation.
(A) Schematic representation of the experimental timeline and setup. (B) Representative images showing ABE8e expression in neurons (white arrowheads) of the visual cortex (+) in the injected hemisphere and the absence of TH+ cell bodies after 12 wk (bottom). Absence of ABE8e expression in neurons (orange arrowheads) of the uninjected hemisphere (top) is shown for comparison (n=3 mice). Scale bars, 20 μm. VC, visual cortex; NEUN, hexaribonucleotide binding protein-3; Cas9, Streptococcus pyogenes Cas9; TH, tyrosine hydroxylase; DAPI, 4’,6-diamidino-2-phenylindole.
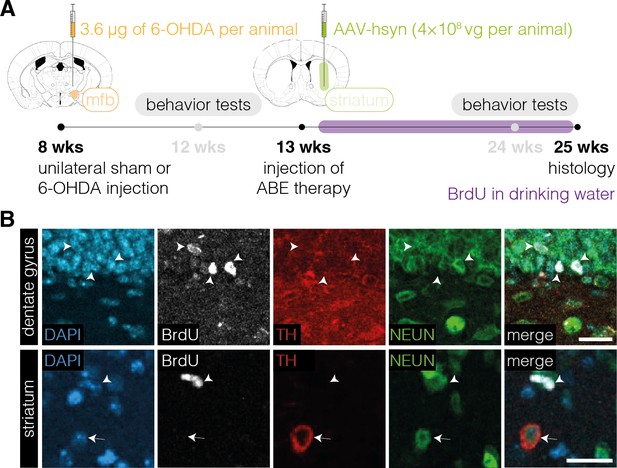
Absence of proliferation in TH+ cell bodies in the striatum after neuronal polypyrimidine tract binding protein 1 (PTBP1) downregulation.
(A) Schematic representation of the experimental timeline and setup. (B) Representative images of BrdU-labeled cells (white arrowhead) in the dentate gyrus (DG, top) or striatum (bottom) and of TH+ cell bodies (white arrow) in the striatum of animals treated with AAV-hsyn (n=4 mice; n=163 TH+ cell bodies in the striatum). Images of the DG are shown as a positive control (top). Scale bars, 20 μm. ABE, adenine base editor; DAPI, 4’,6-diamidino-2-phenylindole; BrdU, bromodeoxyuridine; TH, tyrosine hydroxylase; NEUN, hexaribonucleotide binding protein-3.
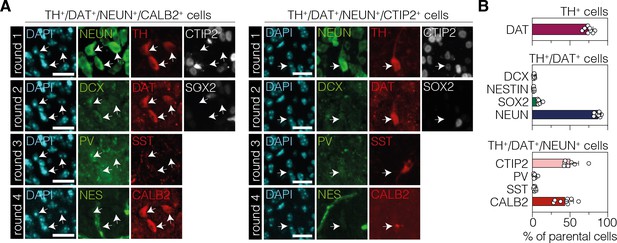
Characterization of TH+ cells in the striatum after neuronal polypyrimidine tract binding protein 1 (PTBP1) downregulation.
(A) Representative 4i images of the two main cell populations among TH-positive cells. The expressed markers are indicated on top of the images. (B) Corresponding quantifications of phenotypic markers expressed among TH-positive cells (top, DAT; n=696 TH+ cells), TH/DAT double-positive (middle, DCX; NESTIN; SOX2; n=527 cells), or TH/DAT/NEUN triple-positive (bottom, CTIP2; PV; SST; CALB2; n=460 cells) subpopulations (white arrows) in the striatum at 12 wk after administration of the AAV-hsyn treatment. The parental population is indicated above each plot. 4i imaging rounds are indicated on the left in (A). Images were pseudocolored during post-processing. Scale bars, 20 μm. Data are displayed as means of six mice (B). DAPI, 4’,6-diamidino-2-phenylindole; SOX2, sex determining region Y-box 2; NES, neuroepithelial stem cell protein; DCX, doublecortin; NEUN, hexaribonucleotide binding protein-3; TH, tyrosine hydroxylase; DAT, dopamine transporter; CTIP2, COUP-TF-interacting protein 2; PV, parvalbumin; SST, somatostatin; CALB2, calbindin 2.
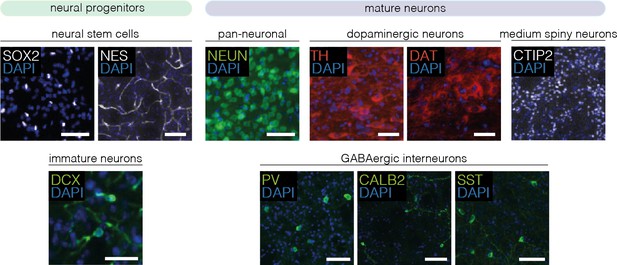
Validation of antibodies for 4i experiments.
Antibody specificity was assessed on mouse brain sections of the striatum or substantia nigra pars compacta (SNc) (n=2 animals). Representative images of one animal are shown. Scale bars, 50 μm. DAPI, 4’,6-diamidino-2-phenylindole; SOX2, sex determining region Y-box 2; NES, neuroepithelial stem cell protein; DCX, doublecortin; NEUN, hexaribonucleotide binding protein-3; TH, tyrosine hydroxylase; DAT, dopamine transporter; CTIP2, COUP-TF-interacting protein 2; PV, parvalbumin; CALB2, calbindin 2; SST, somatostatin.
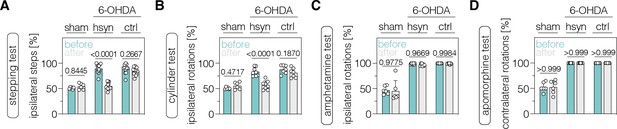
Neuronal polypyrimidine tract binding protein 1 (PTBP1) downregulation in the striatum alleviates drug-free motor dysfunction in 6-OHDA-lesioned Parkinson’s disease (PD) mice.
(A, B) Spontaneous behaviors, were assessed as contralateral forelimb akinesia in the stepping test (A) and spontaneous rotations in the cylinder test (B), in animals treated in the striatum. (C, D) Drug-induced rotations, assessed as amphetamine-induced ipsilateral rotations (C) and apomorphine-induced contralateral rotations (D), in animals treated in the striatum. Normal distribution of the data was analyzed using the Shapiro-Wilk test. Data are represented as means ± s.d. of 6–13 animals per group and were analyzed using a two-way ANOVA with Śidák’s multiple comparisons. Each datapoint represents one animal. Exact p-values are indicated in each plot. hsyn, AAV-hsyn-ABE treatment; ctrl, AAV-ctrl-ABE treatment.
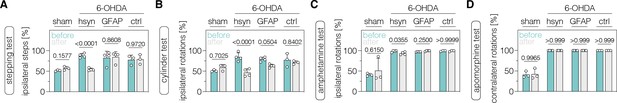
Neuronal polypyrimidine tract binding protein 1 (PTBP1) downregulation in the substantia nigra pars compacta (SNc) alleviates drug-free motor dysfunction in 6-OHDA-lesioned PD mice.
(A, B) Spontaneous behaviors, assessed as contralateral forelimb akinesia in the stepping test (A) and spontaneous rotations in the cylinder test (B), in animals treated in the SNc. (C, D) Drug-induced rotations, assessed as amphetamine-induced ipsilateral rotations (C) and apomorphine-induced contralateral rotations (D), in animals treated in the SNc. Normal distribution of the data was analyzed using the Shapiro-Wilk test. Data are represented as means ± s.d. of 3–4 animals per group and were analyzed using a two-way ANOVA with Śidák’s multiple comparisons test. Each datapoint represents one animal. Exact p-values are indicated in each plot. hsyn, AAV-hsyn-ABE treatment; GFAP, AAV-GFAP-ABE treatment; ctrl, AAV-ctrl-ABE treatment.
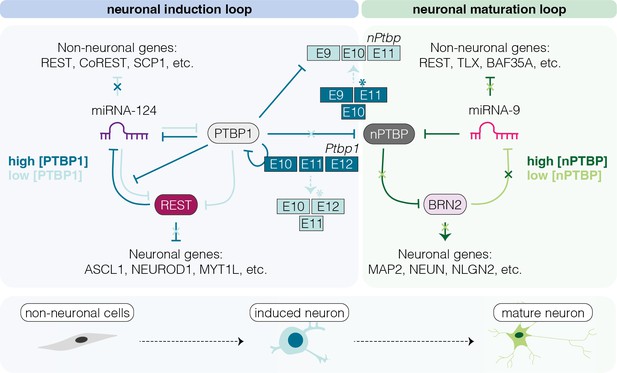
Schematic representation of the polypyrimidine tract binding protein 1 (PTBP1)/nPTBP regulatory loops driving neuronal differentiation and maturation.
The dynamic regulation of PTBP1-mediated inhibition of miRNA-124, which acts on components of the REST complex to activate neuronal-specific expression programs in non-neuronal cells is a key event in the miRNA-124/REST signaling cascade. High PTBP1 expression in non-neuronal cells blocks miRNA-124-induced neuronal differentiation. During neuronal differentiation (low [PTBP1]), miRNA-124 represses PTBP1, enabling expression of nPTBP, pro-neuronal transcription factors, and consequently neuron-specific splicing events that eventually lead to neuronal maturation (Makeyev et al., 2007; Xue et al., 2013).
Additional files
-
Supplementary file 1
List of oligos used for cloning of sgRNA plasmids.
- https://cdn.elifesciences.org/articles/97180/elife-97180-supp1-v1.docx
-
Supplementary file 2
List of oligos used for cloning of adeno-associated virus (AAV) plasmids.
- https://cdn.elifesciences.org/articles/97180/elife-97180-supp2-v1.docx
-
Supplementary file 3
List of oligos used for RT-qPCR.
- https://cdn.elifesciences.org/articles/97180/elife-97180-supp3-v1.docx
-
Supplementary file 4
List of antibodies used in this study.
- https://cdn.elifesciences.org/articles/97180/elife-97180-supp4-v1.docx
-
Supplementary file 5
List of oligos used for deep sequencing.
- https://cdn.elifesciences.org/articles/97180/elife-97180-supp5-v1.docx
-
Supplementary file 6
Reference nucleotide sequences of amplicons for deep sequencing.
- https://cdn.elifesciences.org/articles/97180/elife-97180-supp6-v1.docx
-
Supplementary file 7
Statistic report of the presented data.
- https://cdn.elifesciences.org/articles/97180/elife-97180-supp7-v1.xlsx
-
Supplementary file 8
Summary of GUIDE-seq results in N2a cells.
- https://cdn.elifesciences.org/articles/97180/elife-97180-supp8-v1.xlsx
-
MDAR checklist
- https://cdn.elifesciences.org/articles/97180/elife-97180-mdarchecklist1-v1.docx





