On the role of VP3-PI3P interaction in birnavirus endosomal membrane targeting
Figures
Biophysical characterization of viral protein 3 (VP3) binding to phosphatidylinositol-3-phosphate (PI3P).
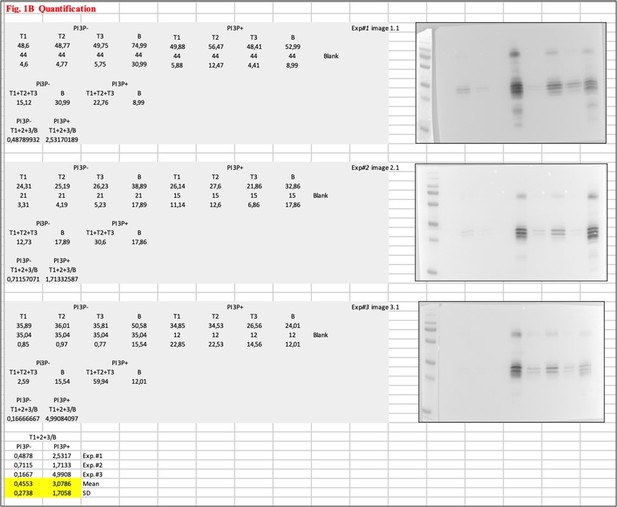
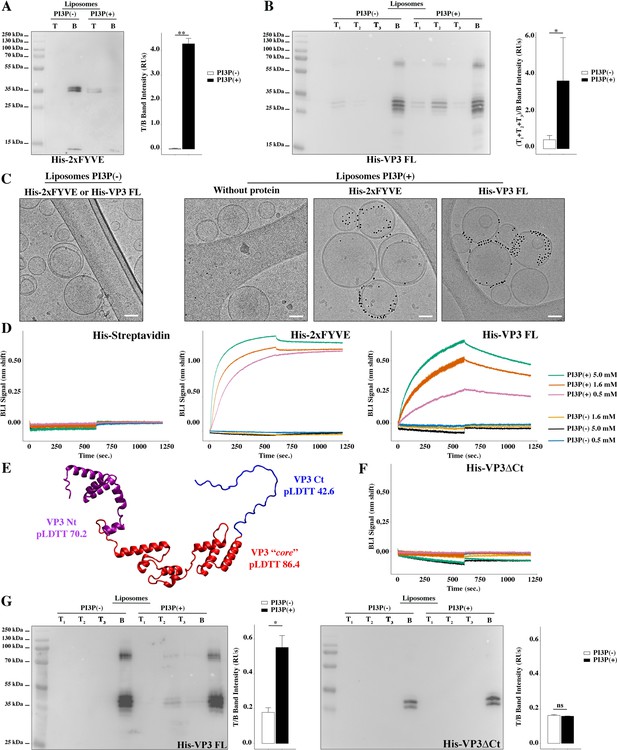
(A) Immunoblots of the top (T) and bottom (B) fractions from a liposome PI3P(-) or PI3P(+) OptiprepTM co-floatation assay indicating that His-2xFYVE protein (~35 kDa) specifically binds to liposome PI3P(+). Results are representative of three independent experiments. The bar plot represents the intensity of T/B bands for each liposome preparation. Significant differences (**p<0.01) as determined by one-way ANOVA with Tukey’s HSD test.(B) Immunoblots of the three top (T1, T2, and T3) and bottom (B) fractions from a liposome PI3P(-) or PI3P(+) OptiprepTM co-floatation assay indicating that His-VP3 FL protein (~32 kDa) specifically binds to liposome PI3P(+). Results are representative of three independent experiments. The bar plot represents the intensity of (T1+T2+T3)/B bands for each liposome preparation. Significant differences (*p<0.05) as determined by one-way ANOVA with Tukey’s HSD test. (C) Liposomes were vitrified and only representative cryo-electron microscopy images are shown. Left panel: liposomes PI3P(-) incubated even with the Ni-NTA gold particles reagent alone or pre-incubated with His-2xFYVE of His-VP3 FL. Right panel: liposomes PI3P(+) control (without protein), or incubated with His-2xFYVE- or His-VP3 FL-Ni-NTA gold particles showing gold particles decorating the membrane of the liposomes when His-2xFYVE or His-VP3 FL were present. The bar represents 50 nm. (D) Binding of His-Streptavidin (negative control), His-2xFYVE (positive control), or His-VP3 FL to three different concentrations of liposomes PI3P(-) or PI3P(+). Association and dissociation sensorgrams measured by bio-layer interferometry (BLI), showing the specific interaction of His-2xFYVE and His-VP3 FL with liposomes PI3P(+) in a dose-dependent manner, as indicated. (E) Cartoon representation of AlphaFold2 prediction of VP3 FL. Red region corresponds to the ‘core’ region of the protein present in the experimental X-ray crystallographic model obtained by Casañas and coworkers PDB: 2R18 (Casañas et al., 2008). Regions not present within the PDB are colored in violet and blue representing the Nt and the Ct of VP3, respectively. pLDTT values lower than 50 are a strong predictor of disorder. (F) Binding of His-VP3 ΔCt to three different concentrations of liposomes PI3P(-) or PI3P(+). Sensorgrams measured by BLI, showing the absence of binding to either liposomes when the VP3 lacks the Ct region blue in (E). (G) Immunoblots of the three top (T1, T2, and T3) and bottom (B) fractions from a liposome PI3P(-) or PI3P(+) OptiprepTM co-floatation assay of His-VP3 FL protein (~32 kDa) (positive control, left panel) or His-VP3 ΔCt protein (~28 kDa) (right panel) showing the lack of VP3 ΔCt binding to both liposomes. Results are representative of three independent experiments. The bar plot represents the intensity of (T1+T2+T3)/B bands for each liposome preparation. Significant differences (*p<0.05; ns p>0.05) as determined by one-way ANOVA with Tukey’s HSD test.
-
Figure 1—source data 1
Original membranes corresponding to Figure 1A, B and G.
- https://cdn.elifesciences.org/articles/97261/elife-97261-fig1-data1-v1.pdf
-
Figure 1—source data 2
Individual files corresponding to the original membranes from Figure 1A, B and G.
- https://cdn.elifesciences.org/articles/97261/elife-97261-fig1-data2-v1.zip
His-viral protein 3 (VP3) Full length (FL) purification and characterization.
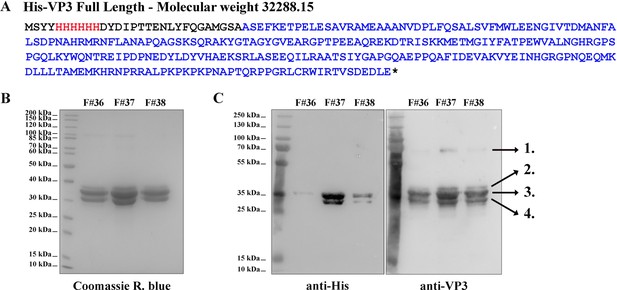
(A) The aminoacidic sequence of His-VP3 FL and its theoretical molecular weight. The His-tag is colored in red and the VP3 sequence in blue. Additional residues are black. (B) Coomassie R. blue-stained polyacrylamide gel showing fractions (F) #36, #37, and #38 from the size exclusion chromatography. (C) Western blot images (left: anti-His; right: anti-VP3) from gels identical to that shown in (B). Numbers 1–4 indicate the identified proteins by mass spectrometry. 1. His-VP3 FL dimer (~70 kDa); 2. Sulfonylated His-VP3 FL (due to the use of phenylmethylsulfonyl fluoride as proteases inhibitor); 3. His-VP3 FL; 4. His-VP3 lacking the last 14 carboxy-terminal (Ct) residues.
-
Figure 1—figure supplement 1—source data 1
Original Coomasie R. Blue-stained polyacrylamide gel and western blot membranes corresponding to Figure 1—figure supplement 1B and C.
- https://cdn.elifesciences.org/articles/97261/elife-97261-fig1-figsupp1-data1-v1.pdf
-
Figure 1—figure supplement 1—source data 2
Individual files corresponding to the original Coomasie R. Blue-stained polyacrylamide gel and western blot membranes from Figure 1—figure supplement 1B and C.
- https://cdn.elifesciences.org/articles/97261/elife-97261-fig1-figsupp1-data2-v1.zip

His-viral protein 3 (VP3) full length (FL) phosphatidylinositol-3-phosphate (PI3P)-binding specificity.
Immunoblots of the three top (T1, T2, and T3) and bottom (B) fractions from co-flotation assays using liposomes where PI3P was replaced by 1,2-dioleoyl-sn-glycero-3-phosphate (PA) or [1,2-dioleoyl-sn-glycero-3-phospho-(1'-myo-inositol)] (PI) in an identical molar ratio, named ‘liposomes PA(+) or PI(+)’. We observed that while His-VP3 FL bound to liposomes PI3P(+) (middle panel), it did not bind to liposomes PA (left panel) or PI (right panel), reinforcing the notion that the interaction of VP3 with PI3P is specific. On top of each panel, the corresponding lipid structure is shown.
-
Figure 1—figure supplement 2—source data 1
Original western blot membranes corresponding to Figure 1—figure supplement 2.
- https://cdn.elifesciences.org/articles/97261/elife-97261-fig1-figsupp2-data1-v1.pdf
-
Figure 1—figure supplement 2—source data 2
Individual files corresponding to the original western blot membranes from Figure 1—figure supplement 2.
- https://cdn.elifesciences.org/articles/97261/elife-97261-fig1-figsupp2-data2-v1.zip
Viral protein 3 (VP3) P2 involvement in the association of VP3 with the early endosomes (EE) membrane.
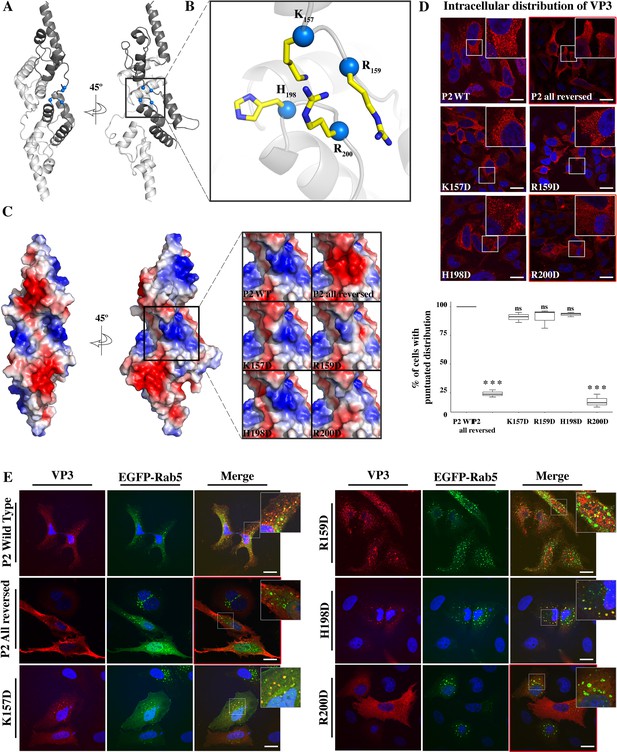
(A) The cartoon representation of the VP3 dimer PDB 2R18 (Casañas et al., 2008) shows each protomer in different shades of gray. The blue balls depict the residues defining the P2 region. (B) The close-up of P2 region showing residues K157, R159, H198,and R200. (C) Electrostatic potential mapped on the surface of the structure of a VP3 dimer in the same orientations as in (A) structures. The close-up shows the impact of P2 residue mutations on the electrostatic potential of the binding site. P2 wild-type (WT) corresponds to the ‘UniProt code’. The color-coded electrostatic surface potential of VP3 was drawn using PyMol (blue positive, red negative). (D) QM7 cells transfected with pcDNA VP3 FL (P2 WT), P2 (all reversed), or the four-point mutants (K157D, R159D, H198D, and R200D) and immunostained with anti-VP3 showing the distribution of each protein (upper panel). Images were captured using a Confocal Laser Scanning Microscopy and then the percentage of cells with punctated fluorescent signal were determined for each protein (lower panel). The red signal shows the VP3 distribution and the blue one shows the nuclei, which were Hoestch-stained. The data were normalized to the P2 WT protein. The box plot represents the percentage of cells with punctuated distribution of VP3. Significant differences (***p<0.001; ns p>0.05.) as determined by one-way ANOVA with Tukey’s HSD test. (E) QM7 cells co-transfected with pEGFP-Rab5 and pcDNA VP3 FL (P2 WT), P2 (all reversed), or the four-point mutants (K157D, R159D, H198D, and R200D) and immunostained with anti-VP3 showing the distribution of each protein. Representative images, captured using a Confocal Laser Scanning Microscopy are shown where green signal represents Rab5 distribution and the red signal that of VP3. Nuclei were Hoestch-stained and are blue. White bar-scales represent 20 mm. VP3 P2 (all reversed) and R200D depict a cytosolic distribution of the proteins. Quantification of the co-localization of the different VP3 proteins and EGFP-Rab5 is shown in Figure 2—figure supplement 1.
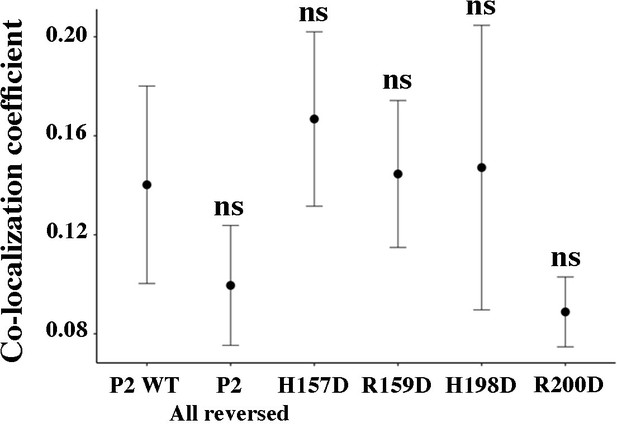
VP3-EGFP-Rab5 co-localization quantification.
The dot plot depicts the co-localization coefficient for each protein determined as explained in the Materials and methods section. Significant differences (ns p>0.05) as determined by one-way ANOVA with Tukey’s HSD test.
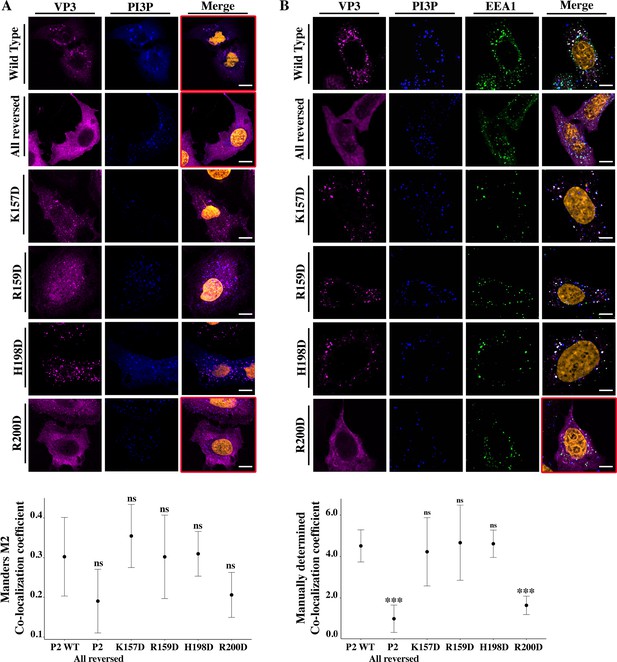
Viral protein 3 (VP3) P2 involvement in the association between VP3 and early endosomes (EE) phosphatidylinositol-3-phosphate (PI3P).
(A) QM7 cells co-transfected with the PI3P biosensor pEGFP-2xFYVE and pcDNA VP3 FL (P2 WT), P2 (all reversed), or the four-point mutants (K157D, R159D, H198D, and R200D) and immunostained with anti-VP3 showing the distribution of each protein. Representative images captured using a Confocal Laser Scanning Microscopy are shown where blue signal represents FYVE distribution and the magenta signal that of VP3. Nuclei were Hoestch-stained and are orange (upper panels). White bar-scales represent 20 mm. VP3 P2 (all reversed) and R200D depict a cytosolic distribution of the proteins with a significant lower co-localization coefficient. The dot plot in the lower panel depicts the co-localization coefficient for each protein determined as explained in the Materials and methods section. Significant differences (ns p>0.05) as determined by one-way ANOVA with Tukey’s HSD test. (B) QM7 cells were transfected with the pcDNA VP3 FL (P2 WT), P2 (all reversed), or the four-point mutants (K157D, R159D, H198D, and R200D). The cells were fixed and then the GST-2xFYVE purified peptide and anti-VP3 antibodies were used to recognize endogenous PI3P and VP3, respectively. Additionally, anti-EEA1 antibodies were used to stain the endosomes. GST-2xFYVE was labelled with a fluorescent anti-GST antibody (blue signal), anti-VP3, and anti-EEA1 antibodies with fluorescent secondary antibodies (magenta and green signals, respectively), and Hoestch-stained nuclei in orange. White bar-scales represent 10 mm. VP3 P2 (all reversed) and R200D depict a cytosolic distribution of the proteins with a significant lower co-localization coefficient. The dot plot in the lower panel depicts the co-localization coefficient for each protein determined as explained in the Materials and methods section. Significant differences (***p<0.001; ns p>0.05) as determined by one-way ANOVA with Tukey’s HSD test.
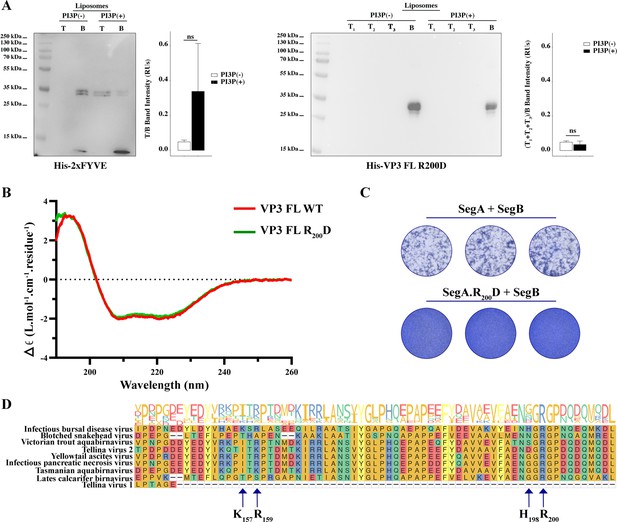
Biophysical characterization of viral protein 3 (VP3) full length (FL) R200D binding to phosphatidylinositol-3-phosphate (PI3P).
(A, left panel). Immunoblots of the top (T) and bottom (B) fractions from a liposome PI3P(-) or PI3P(+) OptiprepTM co-floatation assay indicating that His-2xFYVE protein (~35 kDa) specifically binds to liposome PI3P(+). The bar plot represents the intensity of T/B bands for each liposome preparation. Significant differences (ns p>0.05) as determined by one-way ANOVA with Tukey’s HSD test. (A, right panel). Immunoblots of the three top (T1, T2, and T3) and bottom (B) fractions from a liposome PI3P(-) or PI3P(+) OptiprepTM co-floatation assay indicating that His-VP3 FL R200D protein (~35 kDa) does not bind to liposome PI3P(-) nor PI3P(+). The bar plot represents the intensity of (T1+T2+T3)/B bands for each liposome preparation. Significant differences (ns p>0.05) as determined by one-way ANOVA with Tukey’s HSD test. (B) Far-UV CD spectra of His-VP3 FL (red line) or His-VP3 FL R200D (green line). Spectral acquisitions at 50 nm/min with 0.1 nm steps at 1 s integration time, with a bandwidth of 1 nm were performed four times for the samples as well as for the buffer. The measurements were carried out with constant nitrogen gas flux of 10 ml/min. Acquisitions were averaged and buffer baseline was subtracted with Spectra Manager (JASCO). No smoothing was applied. CDtoolX was used to zero between 255–260 nm and to calibrate the signal amplitude from the fresh CSA signal (Miles and Wallace, 2018). Data are presented as delta epsilon (Δε) per residue (L.mol-1.cm-1.residue1) calculated using the molar concentration of protein and number of residues. (C) QM7 cells were grown in M24 multi-well plate for 12 hr to approximately 90–95% confluency and then 800 ng of plasmids were transfected [(SegA +SegB) or (SegA.R200D+SegB)] in triplicate. At 8 hr post-transfection (p.t.) the supernatants were discarded, and the monolayers were recovered for further plating on M6 multi-well plates containing non transfected QM7 cells. Avicel RC-591 (FMC Biopolymer) was added to the M6 multi-well plates. 72 hr p.i., the monolayers were fixated and stained with Coomassie R250 for revealing the foci forming units. (D) Partial view of amino acid alignment of the VP3 protein for nine reference members of Birnaviridae family. Multiple sequence alignment was performed with Clustal OMEGA (v1.2.4) (Sievers et al., 2011) implemented at EMBL’s European Bioinformatics Institute (Thakur et al., 2024) (The complete alignment is shown in Figure 4—figure supplement 1A). Alignment visualization was done with the R ggmsa package (Zhou et al., 2022) in with assistance from the RStudio software (RStudio Team, 2020). Amino acids are colored according to their side-chain chemistry. Protein sequence logos annotation is displayed on top of the amino acid alignment. For facilitating the view IBDV VP3 142–210 portion is shown. The black arrow indicates the K157, R159, H198, and R200.
-
Figure 4—source data 1
Original membranes corresponding to Figure 4A.
- https://cdn.elifesciences.org/articles/97261/elife-97261-fig4-data1-v1.pdf
-
Figure 4—source data 2
Individual files corresponding to the original membranes from Figure 4A.
- https://cdn.elifesciences.org/articles/97261/elife-97261-fig4-data2-v1.zip

Bioinformatic analysis.
(A) Viral protein 3 (VP3) multiple sequence alignment analysis performed with CLUSTAL Omega (Sievers et al., 2011) with IBDV VP3 (AF_140705.1) as the reference. Characters ‘*,’ ‘:,’ and ‘.’ below the sequences indicate identity (100% of conservation), homology (strongly similar,>50% of conservation), and homology (<50% of conservation), respectively. (B) Matrix showing the percentages of identity of non-IBDV VP3 related to IBDV VP3 (AF_140705.1) performed with CLUSTAL Omega as well. Underlined are indicated those of non-IBDV VP3 related to IBDV VP3.
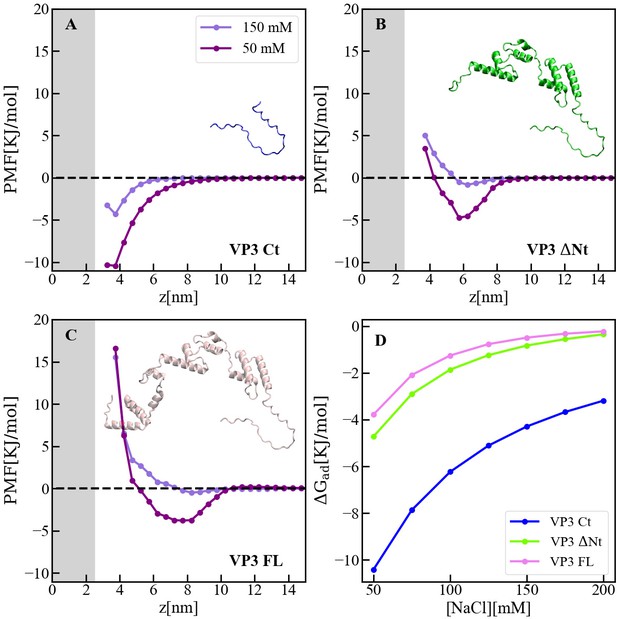
Adsorption of viral protein 3 (VP3) constructs to phosphatidylinositol-3-phosphate (PI3P)(+) model membranes.
(A–C). Adsorption free-energy profiles, PMF(z),for VP3 Ct, VP3 ΔNt, and VP3 FL at 50 and 150 mM of NaCl. (D) Adsorption free energy (ΔGad), computed from the minimum of PMF(z), versus concentration of NaCl. In all cases, the solution pH is 8, and the concentration of protein is 1 µM. The gray areas represent the volume excluded by half the membrane (cis hemilayer, z>0). The membrane surface contains 5% titratable groups, representing PI3P. Each group has three acidic moieties, one with a pKa of 2.5 and the others with 6.5. At 150 mM NaCl and pH 8, more than 90% of the acidic groups are deprotonated (Figure 5—figure supplement 2).
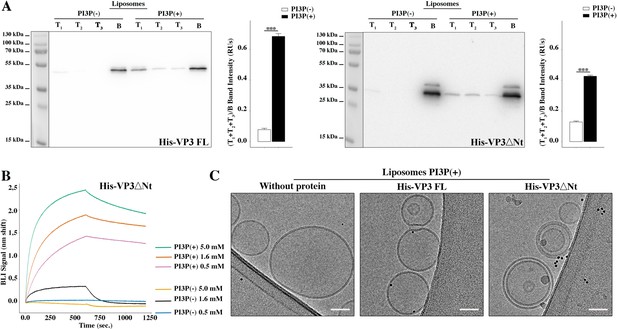
Biophysical characterization of His-viral protein 3 (VP3) DNt binding to phosphatidylinositol-3-phosphate (PI3P).
(A) Left panel: Immunoblots of the three top (T1, T2, and T3) and the bottom (B) fractions from a liposome PI3P(-) or PI3P(+) OptiprepTM co-floatation assay indicating that His-VP3 full length (FL) protein specifically binds to liposome PI3P(+). Results are representative of three independent experiments. The bar plot represents the intensity of (T1+T2+T3)/B bands for each liposome preparation. Right panel: Immunoblots of the three top (T1, T2, and T3) and bottom (B) fractions from a liposome PI3P(-) or PI3P(+) OptiprepTM co-floatation assay indicating that His-VP3 ΔNt protein specifically binds to liposome PI3P(+). Results are representative of three independent experiments. The bar plot represents the intensity of (T1+T2+T2)/B bands for each liposome preparation. Significant differences (***p<0.001) as determined by one-way ANOVA with Tukey’s HSD test. (B) Binding of His-VP3 ΔNt to three different concentrations of liposomes PI3P(-) or PI3P(+). Association and dissociation sensorgrams measured by bio-layer interferometry (BLI), showing the specific interaction of His-VP3ΔNt with liposomes PI3P(+) in a dose-dependent manner, as indicated. (C) Transmission electron microscope images of cryo-fixated liposomes PI3P(+) control (without protein), or incubated with His-VP3 FL- or His-VP3 ΔNt-Ni-NTA gold particles showing electrodense particles decorating the membrane of the liposomes when His-VP3 FL or His-VP3 ΔNt were present. The scale bar represents 50 nm.
-
Figure 5—figure supplement 1—source data 1
Original western blot membranes corresponding to Figure 5—figure supplement 1A.
- https://cdn.elifesciences.org/articles/97261/elife-97261-fig5-figsupp1-data1-v1.pdf
-
Figure 5—figure supplement 1—source data 2
Individual files corresponding to the original western blot membranes from Figure 5—figure supplement 1A.
- https://cdn.elifesciences.org/articles/97261/elife-97261-fig5-figsupp1-data2-v1.zip
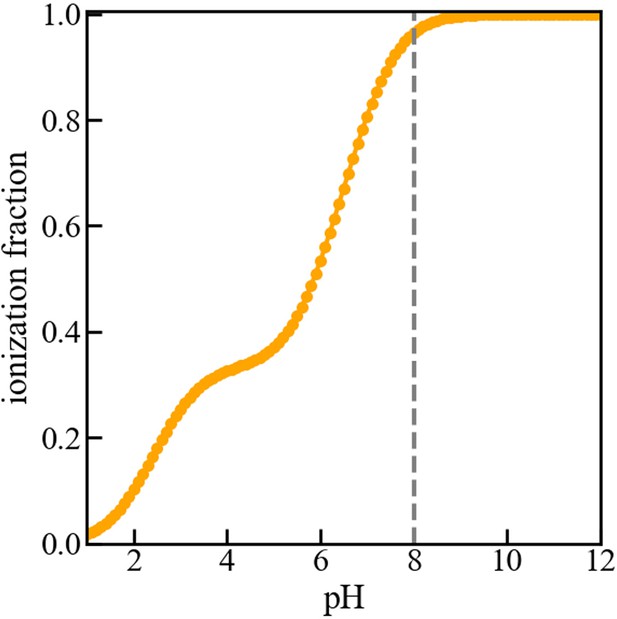
Titration curve of the membrane.
Fraction of deprotonated (or ionized) acidic groups on the surface of the membrane versus pH of the bulk solution. The dashed vertical line indicates that at pH 8, the acidic groups are nearly fully deprotonated, resulting in a negative surface charge on the membrane. As mentioned in the manuscript, the model membrane consists of a 5 nm wide slab of dielectric material that exposes a mixture of 5% acidic and 95% neutral ‘head groups’ to a 150 mM NaCl solution. The acidic lipids contain three acid moieties with pKa values of 2.5 and 6.5, as determined for phosphatidylinositol-3-phosphate (PI3P). No proteins are present in the solution for the purpose of computing the titration curve. The curve was obtained using the Molecular Theory briefly described in the manuscript and fully detailed in the references (Chiarpotti et al., 2021; Ramírez et al., 2019).
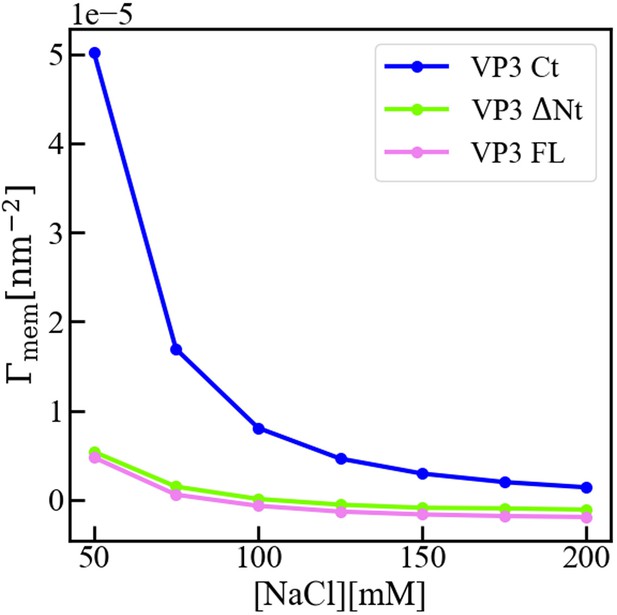
Interfacial concentration of viral protein 3 (VP3) constructs.
Molecular Theory calculation of the surface concentration, or more precisely the surface excess concentration of the three protein constructs VP3 carboxy-terminal (Ct), VP3 ΔNt, and VP3 full length (FL), as a function of the concentration of salt in the bulk solution. The overall electrical charge of each construct is +5, 0, and –3, respectively. The pH of the solution is 8, so that the surface of the membrane is negatively charged. The surface concentration of each protein decreases when increasing the concentration of salt, as the salt ions screen the electrostatic interactions between the proteins and the surface of the membrane. Clearly, the effect is stronger for positively charged VP3 Ct. As detailed in reference (Chiarpotti et al., 2021) the surface excess is defined as where is the average protein density at position z, and is that in the bulk solution. gives the total number of proteins per unit area, in excess of the bulk solution, that accumulate () or deplete () due to the presence of the membrane.
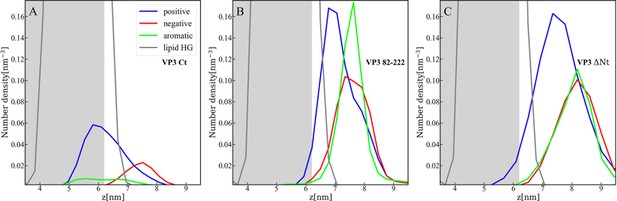
Distribution of charged and aromatic residues of viral protein 3 (VP3).
Spatial distribution of charged groups obtained from coarse-grained Molecular Dynamics simulations of VP3 carboxy-terminal (Ct) (A), VP3 82-222 (B), and VP3 ΔNt (C) adsorbed on a lipid bilayer composed of 1,2-dioleoyl-sn-glycero-3-phosphoethanolamine (DOPE), 1,2-dioleoyl-sn-glycero-3-phosphocholine (DOPC), and phosphatidylinositol-3-phosphate (PI3P) (with molar ratios of 64:31:5). DOPE and DOPC are electroneutral, while PI3P is anionic. VP3 82-222 represents a portion of the VP3 sequence for which a crystallographic structure is known (Gly82-Asn222). It was included in the plot for comparison with VP3 Ct (Arg223-Glu257) and VP3 ΔNt (Gly82-Glu257). The variable 'z' represents the z-component of the perpendicular distance between the center of mass of the membrane and the center of mass of each protein. The blue curves represent the distribution of positively charged residues in the protein, while the red curves represent the distribution of negatively charged residues. The green curves depict the distribution of aromatic residues, and the gray curves represent the distribution of the lipid’s polar head groups. The gray shaded area schematically represents the region predominantly occupied by the lipids. As observed, in all cases, the positively charged amino-acids are in closer contact with the membrane surface and penetrate into the region of the lipid’s polar head groups. In VP3 ΔNt, negatively charged and aromatic residues are somewhat more exposed towards the solution side of the interface.
Map of electrostatic potential around viral protein 3 (VP3) full length (FL).
Four different views of the electrostatic potential map around VP3 FL, as computed with the Adaptive Poisson-Boltzmann Solver (APBS) plugin of VMD (Dolinsky et al., 2004). Electrostatic isopotential surfaces at +1.1 mV in red and at –1.1mV in blue account for negative and positive electrostatic potential, respectively.

Viral protein 3 (VP3) approaching a lipid bilayer, and distortion of the membrane.
(A–D) Temporal sequence of configurations illustrating how VP3 ΔNt approaches the negatively charged membrane during a 500 ns molecular dynamics (MD) simulation. The membrane contains 1,2-dioleoyl-sn-glycero-3-phosphoethanolamine (DOPE),1,2-dioleoyl-sn-glycero-3-phosphocholine (DOPC), and phosphatidylinositol-3-phosphate (PI3P) in 64:31:5 molar ratio. The beads making the carboxy-terminal (Ct) fragment are colored blue. (E) Magnification of the protein configuration bound to the membrane, with P2 residues represented in cyan balls. (F) Radial distribution function, g(r), between the center of mass of VP3 ΔNt and the center of PI3P molecules in the cis hemilayer. g(r) greater than 1 implies local enhancement of PI3P concentration. The inset shows an upper view of VP3 ΔNt bound to the membrane, with the area within r=4 nm colored in orange. (G) Cryo-electron microscopy images of cryo-fixated liposomes PI3P(+) control (without protein, left panel), or incubated with His-VP3 FL showing the small pinches or localized thinnings in the bilayer of the liposomes when His-VP3 FL was present (middle panel). The bar represents 50 nm. The right panel represents an enlarged image of the red square on the middle panel. White arrows point to the small pinches or localized thinnings in the bilayer when His-VP3 FL was present.
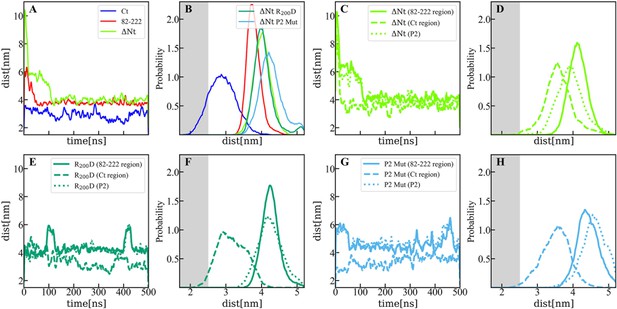
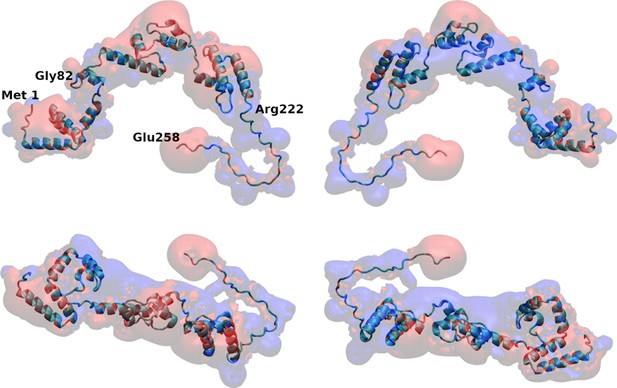
Distance between membrane and protein constructs computed by molecular dynamics (MD).
(A) Time evolution of the distance (perpendicular to the membrane surface) between the center of mass of viral protein 3 (VP3) carboxy-terminal (Ct) (blue solid line), VP3 82–222 (red solid line), and VP3 DNt (green solid line) and the center of mass of the membrane. (B) Distribution of distances computed from the time-traces of panel A in the time interval [200-500] ns; and also from simulations of two mutants of VP3 ΔNt: the single mutant R200D (VP3 ΔNt R200D) and the one with the four residues of the P2 region mutated K157D, R159D, H198D and R200D (VP3 ΔNt P2 Mut). The VP3 Ct construct gets closer to the membrane than any other construct or protein domain and, more importantly, VP3 ΔNt adsorbs with the Ct region facing the negatively charged membrane. The single mutant exhibits a binding similar to wild-type VP3 ΔNt protein, while the one with the whole P2 mutated is located further away from the negative surface. Results obtained by coarse-grained Molecular Dynamics simulation of VP3 Ct, VP3 82–222, and VP3 ΔNt, in front of a 1,2-dioleoyl-sn-glycero-3-phosphoethanolamine (DOPE), 1,2-dioleoyl-sn-glycero-3-phosphocholine (DOPC), phosphatidylinositol-3-phosphate (PI3P) membrane (molar ratios of 64:31:5). (C, E, G) Time evolution of the distance between the center of mass of different protein regions and the center of mass of the membrane for VP3 ΔNt and the two mutants. The continuous line corresponds to the center of mass of the region 82–222, the dashed lines, to the Ct region, and the dotted lines, to the P2 region.(D, F, H) Distribution of distances computed from the time-traces of panels C, E, and G in the time interval [200- 500] ns.
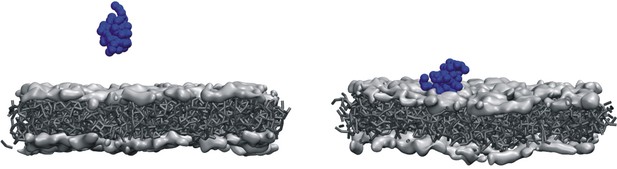
Initial and final configurations of viral protein 3 (VP3) carboxy-terminal (Ct) on a lipid membrane.
Initial (left panel) and final (right panel) configurations from a 500 ns coarse-grained Molecular Dynamics simulation of VP3 Ct in front of a 1,2-dioleoyl-sn-glycero-3-phosphoethanolamine (DOPE), 1,2-dioleoyl-sn-glycero-3-phosphocholine (DOPC), phosphatidylinositol-3-phosphate (PI3P) membrane (molar ratios of 64:31:5).
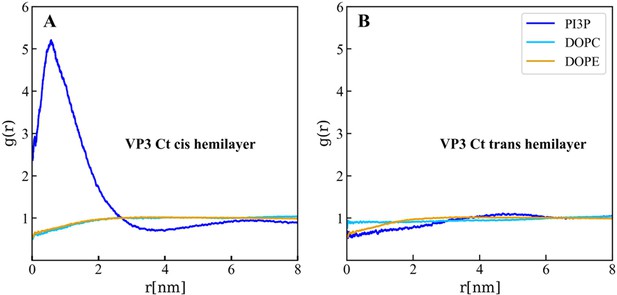
Lipids accumulation/depletion around viral protein 3 (VP3) carboxy-terminal (Ct).
In plane (X–Y) radial distribution functions, g(r). These functions were computed during the 200–500 ns time interval of the Molecular Dynamics simulations described in the text and in the previous supporting figures. The panels display the g(r) between the lipids in the cis- (left) and trans- (right) membrane hemilayers, and the center of mass of the VP3 Ct construct. Once again, this positively charged construct attracts the PI3P molecules from the cis-hemilayer to its proximity, as evident from the values of g(r)>1 for r<2 nm. Moreover, for VP3 Ct this effect is strong enough to deplete the PI3P molecules in the trans-hemilayer due to inter-hemilayer phosphatidylinositol-3-phosphate (PI3P)-PI3P electrostatic repulsions.
Lipids accumulation/depletion around the carboxy-terminal (Ct) fragment of viral protein 3 (VP3) DNt.
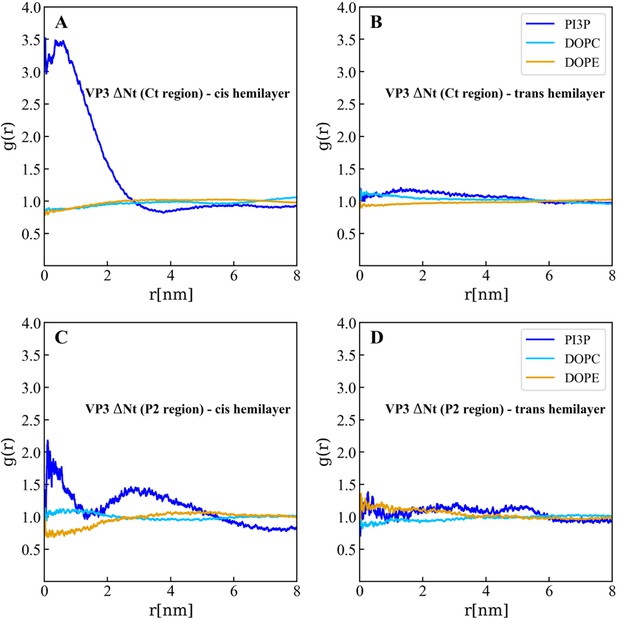
In plane (X–Y) radial distribution functions, g(r). These functions were computed during the 200-500 ns time interval of the Molecular Dynamics simulations described in the text and in the previous supporting figures. The upper panels display the g(r) between the lipids in the cis- (left) and trans- (right) membrane hemilayers, and the center of the Ct domain of VP3 ΔNt. As observed, the Ct domain attracts phosphatidylinositol-3-phosphate (PI3P) to its proximity, within a radius of approximately 4 nm where g(r)>1 (blue line), in the hemilayer that is in contact with the protein (cis-). The lower panels depict the radial distribution functions between the lipids in the cis- (left) and trans- (right) hemilayers, and the P2 domain of VP3 ΔNt. Once again, P2 attracts PI3P to its proximity in the cis-hemilayer, within a radius of about 5 nm.
Lipids accumulation/depletion around viral protein 3 (VP3) ΔNt and its mutants.
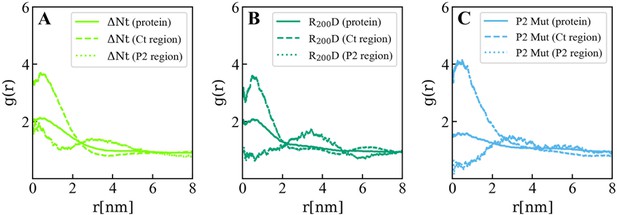
In plane (X–Y) radial distribution functions, g(r) computed in the 200-500 ns time interval of the Molecular Dynamics simulations of the VP3 ΔNt protein (A) and two mutants: VP3 ΔNt R200D (B) and VP3 ΔNt P2 Mut (C). The panels display the g(r) between phosphatidylinositol-3-phosphate (PI3P) lipids in the cis membrane hemilayer, and the center of mass of the whole protein (continuous line), the Ct region (dashed line), and the P2 region (dotted line). The concentration effects on PI3P, induced by VP3 ΔNt, is exerted by the positively charged Ct and the P2 regions. When the P2 region is mutated, the P2 contribution to PI3P recruitment is diminished gradually from the single mutant to the wholly mutated P2.
Adsorption free energy of viral protein 3 (VP3) ΔNt and VP3 ΔNt mutants on PI3P(+) model membranes.
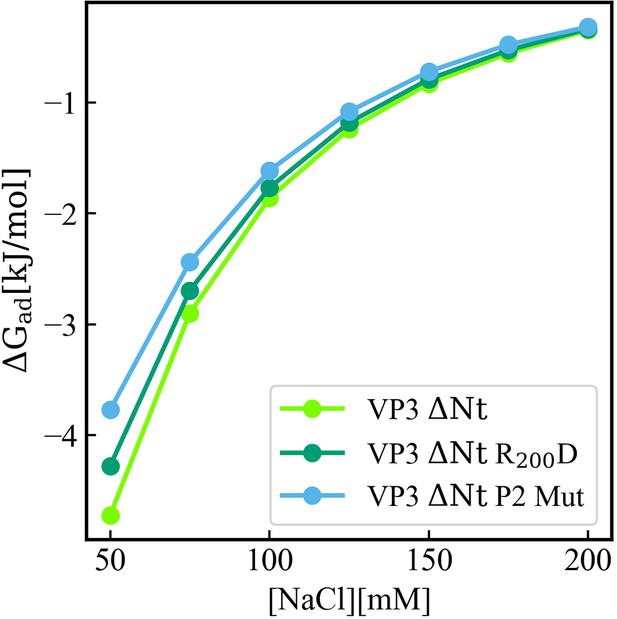
Adsorption free energy (ΔGad), computed from the minimum of PMF(z), versus concentration of NaCl, for VP3 ΔNt and the mutants VP3 ΔNt R200D and VP3 ΔNt P2 Mut. In all cases, the solution pH is 8, and the concentration of protein is 1 µM. The membrane surface contains 5% titratable groups, representing phosphatidylinositol-3-phosphate (PI3P). Each group has three acidic moieties, one with a pKa of 2.5 and the others with 6.5. At 150 mM NaCl and pH 8, more than 90% of the acidic groups are deprotonated. As the number of positively charged residues in VP3 is systematically reduced from VP3 ΔNt to VP3 ΔNt P2 Mut, the binding of the protein to the membrane becomes weaker. The effect is more pronounced at lower salt concentrations, which highlights the weight of electrostatic forces on the adsorption of VP3 on negatively charged membranes.
Additional files
-
Supplementary file 1
Primers.
All the primers used for this work. * For the first four-point mutants, the nucleotides that allow to introduce amino acid changes in the VP3 protein are indicated in italic bold letters. The underlined nucleotides indicate restriction sites.
- https://cdn.elifesciences.org/articles/97261/elife-97261-supp1-v1.docx
-
MDAR checklist
- https://cdn.elifesciences.org/articles/97261/elife-97261-mdarchecklist1-v1.docx