The PRC2.1 subcomplex opposes G1 progression through regulation of CCND1 and CCND2
Figures
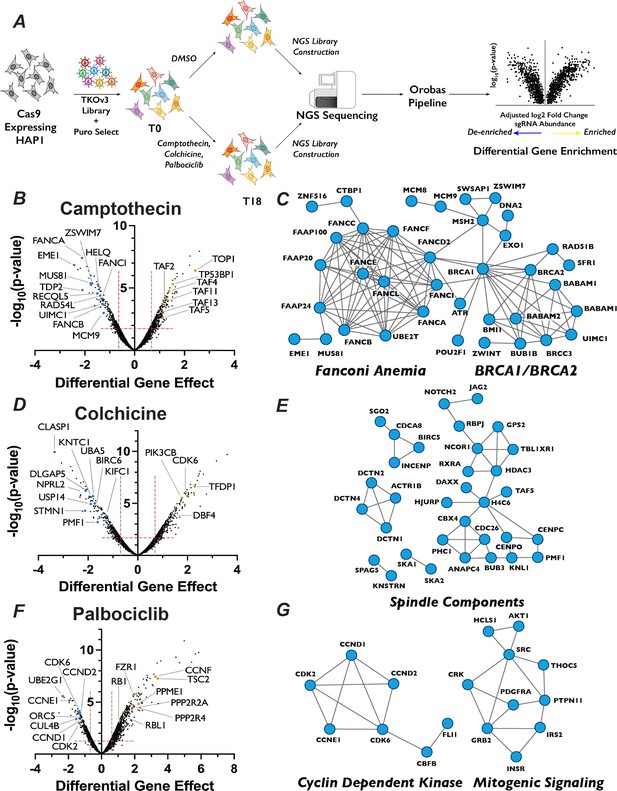
Chemogenetic CRISPR–Cas9 screen to study cell cycle progression.
(A) Schematic of chemogenetic CRISPR–Cas9 screen. (B) Volcano plots of camptothecin chemogenetic screen results. The ‘Differential Gene Effect’ was plotted against the −log10(p-value) for this effect for each gene targeted in the screen, as calculated by the Orobas pipeline. Red dotted line indicates the established cut-off. Highlighted dots are genes with known roles in response to each treatment, with blue or yellow dots indicate genes that when inactivated resulted in sensitivity or resistance, respectively, to camptothecin. (C) Representative STRING analysis networks for protein complexes with known roles in pathways that we identified as sensitive in the camptothecin chemogenetic screen. Blue dots in the STRING network indicate genes that when inactivated resulted in sensitivity to camptothecin. (D) Same as in (B) but for colchicine chemogenetic screen results. (E) Same as in (C) but for colchicine screen results. (F) Same as in (B) but for palbociclib chemogenetic screen results. (G) Same as in (C) but for palbociclib screen results.
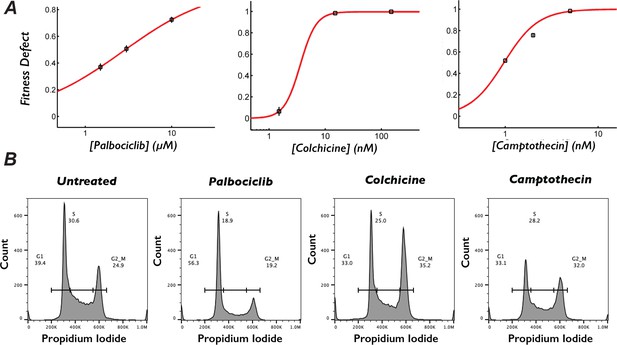
Dosing to determine inhibitor concentration for chemogenetic screens.
(A) Drug dosing experiments were performed to determine screening concentrations. Cells were counted during passage in increasing doses of camptothecin (left), palbociclib (center), and colchicine (right). (B) Representative images of flow cytometry traces from untreated cells or cells treated with 0.7 µM palbociclib-, 9.2 nM colchicine-, or 1 nM camptothecin-treated cells for 3 days, then stained propidium iodide. Plots represent the number of stained cells with a given propidium iodide intensity.
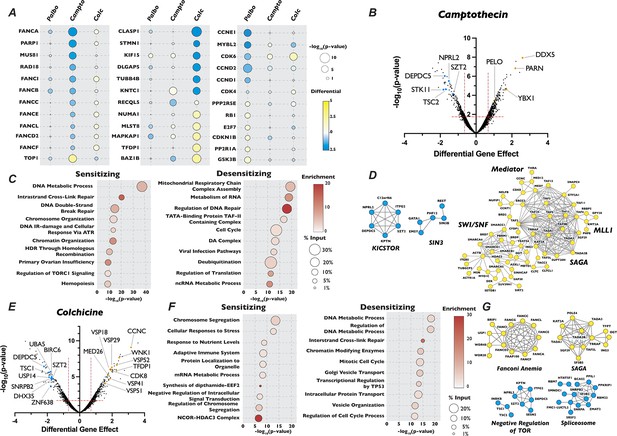
Analysis of camptothecin and colchicine chemogenetic screen reveals novel players in cell cycle regulation.
(A) Dot plot comparison of the effect of gene mutation across three different screen conditions. Circle color indicates the strength of the positive or negative differential gene effect, circle size indicates the −log10(p-value) of the sgRNA enrichment. (B) Volcano plot of genes identified in the camptothecin chemogenetic screen, plotted as in Figure 1B with highlighted dots representing novel genes identified in the camptothecin screen. (C) Dot plot of Metascape analysis of significant genes that sensitized or de-sensitized cells to camptothecin. The −log10(p-value) of each term was plotted the enrichment was indicated by color of circle and the percentage of the input of genes associated with a given term is indicated by the size of the circle. (D) STRING analysis of genes identified from the analysis of the camptothecin screen. (E), (F), and (G) Same as in (B), (C), and (D) except for the colchicine screen.
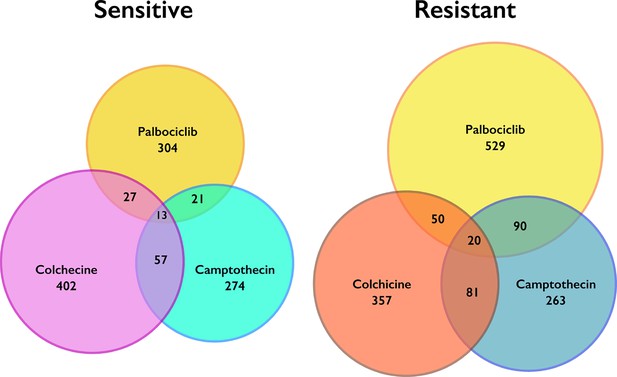
Unique and shared genes identified as significantly enriched or de-enriched in chemogenetic screens.
Venn diagrams showing overlap for significant genes that sensitized (left) or de-sensitized cells (right) to each condition tested. Genes that were determined as significant in all three screens were omitted in further analyses.
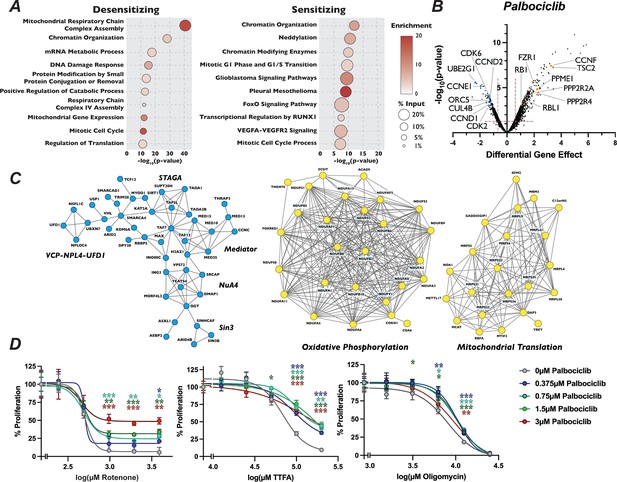
Mutation of mitochondria genes attenuates the sensitivity to palbociclib.
(A) Dot plot of the −log10(p-value) Metascape analysis of significant genes in the palbociclib chemogenetic screen. The enrichment of a given term is indicated by color of circle and the percentage of the input is indicated by the size of the circle. (B) Volcano plot of genes identified from our analysis of the palbociclib screen, plotted as in Figure 1D, with highlighted dots representing novel genes. (C) STRING networks of novel protein complexes identified in palbociclib screen. Dots in the STRING network indicate genes that when inactivated resulted in sensitivity (blue) or resistance (yellow) to palbociclib. (D) Dose–response curve of palbociclib-induced proliferation rescue in combination with oxidative phosphorylation inhibitors by PrestoBlue assay. Cells were grown in palbociclib with or without increasing concentrations of rotenone, thenoyltrifluoroacetone (TTFA), or oligomycin. Data represent mean of three technical replicates, normalized to the initial dose of each inhibitor in indicated concentration of palbociclib, ±StdDev. *p-value <0.05, **p-value <0.005, ***p-value <0.0005, n.s.: not significant, two-tailed unpaired Student’s t-test.
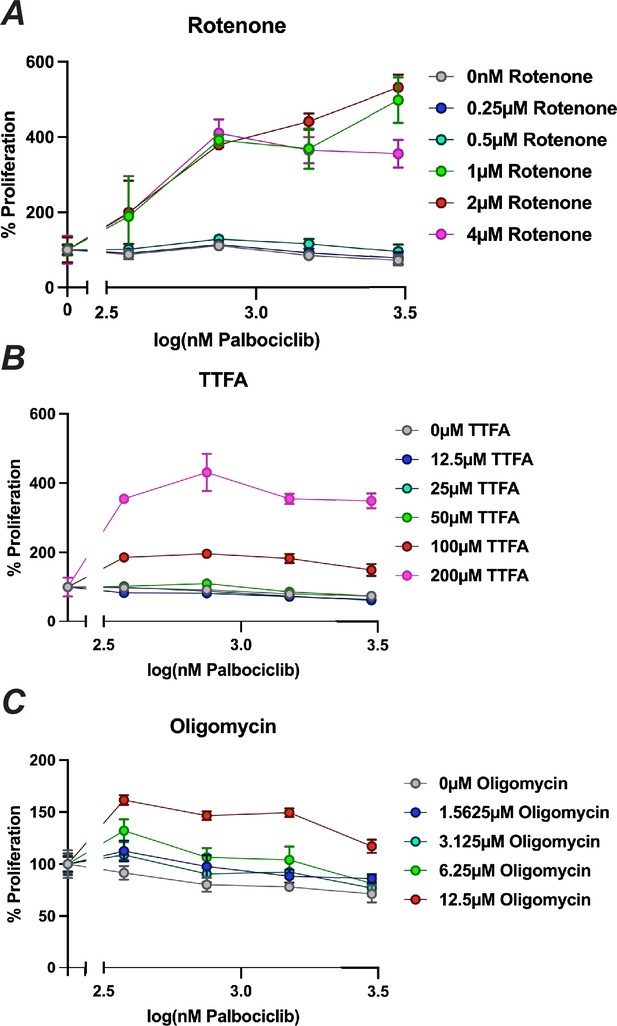
Alternative representation of dose–response curves for combination of oxidative phosphorylation and palbociclib.
(A) Dose–response curve of palbociclib-induced proliferation rescue in combination with oxidative phosphorylation inhibitors by PrestoBlue assay. Data represent mean of three technical replicates, normalized to the initial dose of each inhibitor in indicated concentration of rotenone, ±StdDev. (B) Same as in (A) but for TTFA. (C) Same as in (A) but for oligomycin.
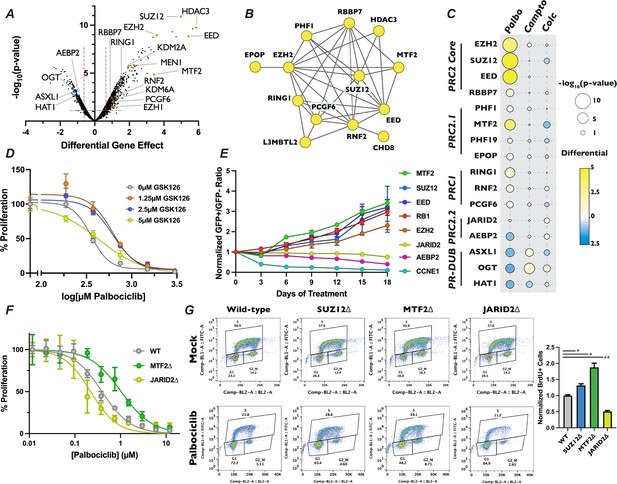
Loss of polycomb repressive complex components display specific resistance to palbociclib.
(A) Volcano plot as in Figure 3B except with members of PR-DUB, PRC1, and PRC2 highlighted. (B) STRING analysis network of PRC components. Yellow dots indicate that inactivation of these genes conferred resistance to palbociclib. (C) Dot plot of comparison of the effect of PRC2 complex member gene mutation across three different screen conditions, as in Figure 2B. (D) Dose–response curve of palbociclib-induced proliferation inhibition rescue with GSK126 by Crystal Violet assay. Data were normalized to untreated cells and represents the mean of three technical replicates, ±StdDev. (E) Results of competitive proliferation assay for each indicated time point, normalized to the initial GFP+/GFP− ratio of the pool. The performance of each sgRNA in 1.5 µM palbociclib versus Mock is shown, after normalizing to control sgRNAs, ± d Graduate Program, University of CalifSEM of the GFP+/GFP− ratios of three independent sgRNAs. (F) Dose–response curve of palbociclib-induced proliferation inhibition in MTF2∆ and JARID2∆ cells by Crystal Violet assay. Data represents mean staining of three monoclonal knockout cell lines, ±StdDev. (G) BrdU incorporation assay for wild-type, SUZ12∆, MTF2∆, and JARID2∆ cell lines. Left – Representative BrdU incorporation versus propidium iodide flow cytometry traces. Right – Quantification of BrdU incorporation assay, mean of S-phase cells in three knockout lines ±StDev. *p-value <0.05, **p-value <0.005, n.s.: not significant, two-tailed unpaired Student’s t-test.
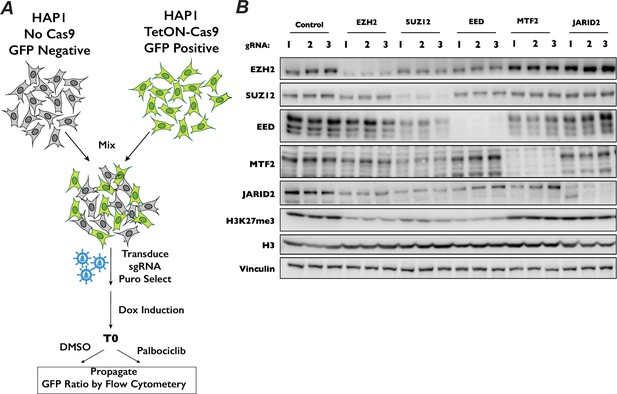
Competitive proliferation assay to determine resistance of PRC2 component mutants to CDK4/6 inhibitors.
(A) Schematic of internally controlled competitive proliferation assay used to validate chemogenetic results or knockout cell line proliferation when treated with palbociclib. In experiments where we generated pooled knockouts, GFP+ cells expressing Cas9 were mixed with GFP− cells without Cas9 (as in Figure 4E). For competitive proliferation experiments with monoclonal knockout cell lines, GFP+, Cas9-expressing cells were mixed with GFP− monoclonal knockout lines (as was done in Figure 4—figure supplement 2). (B) Western blots demonstrating the efficacy of indicated sgRNA used in the competitive proliferation assay.
-
Figure 4—figure supplement 1—source data 1
Original files for western blots shown in Figure 4—figure supplement 1B.
- https://cdn.elifesciences.org/articles/97577/elife-97577-fig4-figsupp1-data1-v1.zip
-
Figure 4—figure supplement 1—source data 2
Original files for western blots shown in Figure 4—figure supplement 1B, indicating relevant band.
- https://cdn.elifesciences.org/articles/97577/elife-97577-fig4-figsupp1-data2-v1.zip
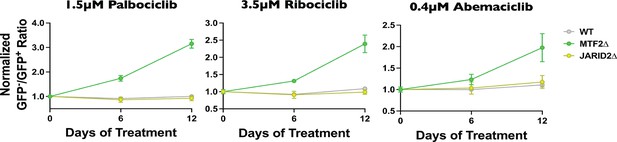
MTF2∆ mutant cell lines display resistance CDK4/6 inhibitors in competitive proliferation assay.
Wild-type, MTF2∆, and JARID2∆ cell lines (GFP−) were mixed with wild-type cells expressing Cas9 and GFP (GFP+) and treated with either DMSO (mock) or 1.5 µM palbociclib (left), 3.5 µM ribociclib (center), or 0.4 µM abemaciclib (right). Cells were split every 3 days and the GFP−/GFP+ ratio was assessed every 6 days by flow cytometry.

PARP cleavage does not change in MTF2∆ or JARID2∆ knockout mutant cell lines upon palbociclib treatment.
Western blot of protein extracts from cells treated with DMSO (mock) or 1.5 µM palbociclib for 48 hr, probed with indicated antibody. PARP cleavage and BIM from protein extracts from RPE1 cells over-expressing a doxycycline-inducible HA-tagged BIM to induce apoptosis as a control.
-
Figure 4—figure supplement 3—source data 1
Original files for western blots shown in Figure 4—figure supplement 3.
- https://cdn.elifesciences.org/articles/97577/elife-97577-fig4-figsupp3-data1-v1.zip
-
Figure 4—figure supplement 3—source data 2
Original files for western blots shown in Figure 4—figure supplement 3, indicating relevant band.
- https://cdn.elifesciences.org/articles/97577/elife-97577-fig4-figsupp3-data2-v1.zip
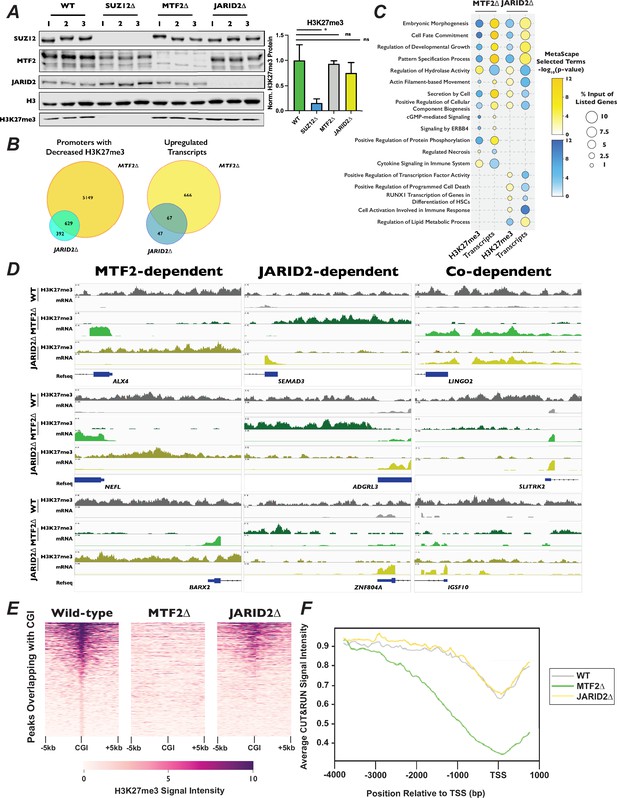
Polycomb 2.1 and PRC2.2 are differentially recruited to promoters with CpG islands.
(A) Left – Western blots of wild-type, SUZ12∆, MTF2∆, and JARID2∆ cell extracts probed with the indicated antibodies.Right – Quantification of H3K27me3 signal intensity, normalized to H3, ±StDev. *p-value <0.05, n.s.: not significant, two-tailed unpaired Student’s t-test. (B) Venn diagrams of MTF2∆ or JARID2∆ compared to wild-type cells of left – promoters with decreased H3K27me3 signal in CUT&RUN experiment or right – increased transcript levels in RNA-Seq. (C) Dot-plot of selected Metascape terms of protein-coding genes displaying significantly increased or decreased levels of H3K27me3 or transcripts. Color of the circle indicates the −log10(p-value) of the term and the size of circle indicates the percentage of the genes from the input list were represented in that term. (D) Genome browser traces of promoters with decreased H3K27me3 and increased mRNAs that were dependent on MTF2 (left), JARID2 (center), or on the presence either MTF2 or JARID2 (right). Tracks represent combined BED files from two clonal biological replicates. (E) Representative heatmap of H3K27me3 signal for 1877 peaks overlapping with CGI. Genomic regions are ordered by the H3K27me3 read density intensity in wild-type cells then plotted for the same loci in MTF2∆ and JARID2∆ cells. Plots are of one of two biological replicates. (F) H3K27me3 signal averaged for all CGI-containing promoters for wild-type, MTF2∆, and JARID2∆ cells.
-
Figure 5—source data 1
Original files for western blots shown in Figure 5A.
- https://cdn.elifesciences.org/articles/97577/elife-97577-fig5-data1-v1.zip
-
Figure 5—source data 2
Original files for western blots shown in Figure 5A, indicating relevant band.
- https://cdn.elifesciences.org/articles/97577/elife-97577-fig5-data2-v1.zip
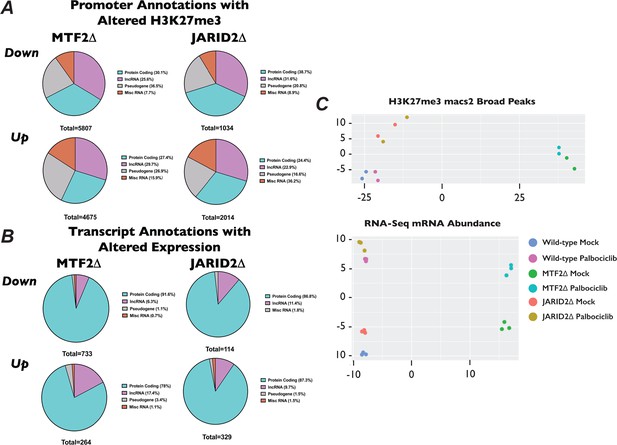
Changes in H3K27me3 distribution and differentially expressed genes in MTF2∆ and JARID2∆ cells.
(A) Venn diagrams of the Gencode Annotations of promoters that had significantly upregulated (top row) and downregulated H3K27me3 (bottom row) for MTF2∆ (left) and JARID2∆ cells (right). Significant promoters were determined as having a log2 fold-change ±1 and an adjusted p-value of <0.1. (B) Same as in (A) only for our RNA-Seq experiments and significant promoters were determined as having a log2 fold-change ±1 and an adjusted p-value <0.05. (C) Top – Primary component analysis (PCA) plot of H3K27me3 peaks called by macs2 from CUT&RUN experiment done in biological duplicate. Bottom – PCA plot of RNA-Seq reads for experiment in biological triplicate.
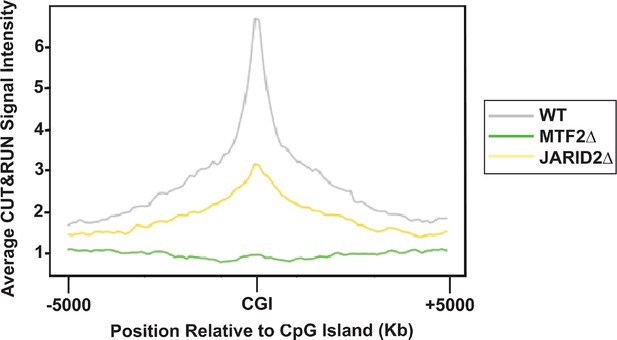
Average H3K27me3 distribution over a 10 kb window for 1877 peaks overlapping with CGIs in MTF2∆ and JARID2∆ cells.
Genomic regions are ordered by the H3K27me3 read density intensity in wild-type cells then plotted for the same loci in MTF2∆ and JARID2∆ cells. Plots are of one of two biological replicates.
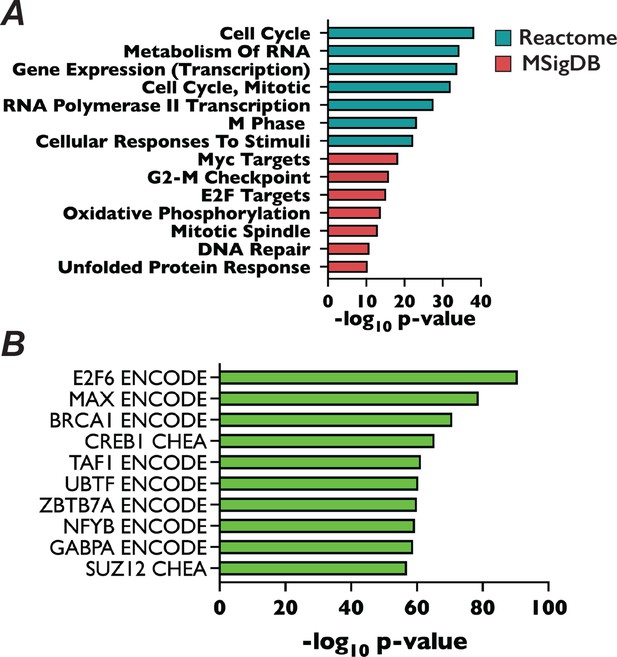
Functional analysis of CGIs.
(A) Bar plot of log10(p-value) of Reactome (teal bars) and MSigDB (red bars) terms associated with promoters of protein-coding genes that contain at least one CGI. (B) Bar plot of −log10(p-value) for the enrichment of a given transcription factors from ENCODE and ChEA databases binding to the list of promoters with overlapping GGIs and H3K27me3 peaks.
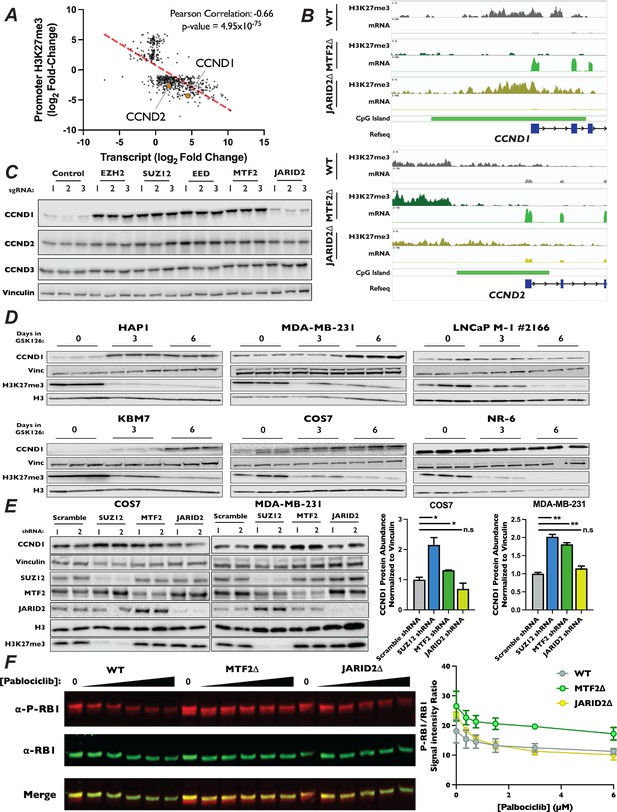
CCND1 and CCND2 expression is increased in MTF2∆ mutants.
(A) Scatterplot of genes whose log2 fold-changes for MTF2∆/wild-type ratio of mRNA expression (x-axis) versus promoter H3K27me3 signal (y-axis) had an adjusted p-value of <0.05 and an adjusted p-value <0.1 where plotted. (B) Genome browser traces of H3K27me3 signal, transcript abundance and CGIs in the CCND1 and CCND2 promoters. Annotated CGIs indicated by green bar. (C) Top – Western blots of Cas9-expressing pools of cells transduced three independent sgRNAs targeting the indicated genes, probed with the indicated antibodies. (D) Western blots of whole-cell lysates from a panel of cell lines treated with 10 µM GSK126 for the indicated time points, with listed antibodies. (E) Left – Western blots of whole-cell lysates from MDA-MB-231 and COS7 cells transduced with shRNA constructs shRNAs targeting SUZ12, MTF2, JARID2, or a scrambled control. Probed with indicated antibodies. Right – Quantification of western blots, CCND1 signal normalized to Vinculin. Each bar is the mean for two different shRNA expressing pools, error bars ±range. *p-value <0.05, **p-value <0.005, n.s.: not significant, two-tailed unpaired Student’s t-test. (F) Left – Representative western blot of total RB1 and P-S807/8111-RB1 with increasing [palbociclib] in WT, MTF2∆, and JARID2∆ cells, probed with indicated antibodies. Right – Quantification of the ratio of P-S807/8111-RB1 to total RB1 signal plotted against [palbociclib], two biological replicates, error bars ±range.
-
Figure 6—source data 1
Original files for western blots shown in Figure 6C–F.
- https://cdn.elifesciences.org/articles/97577/elife-97577-fig6-data1-v1.zip
-
Figure 6—source data 2
Original files for western blots shown in Figure 6C–F, indicating relevant band.
- https://cdn.elifesciences.org/articles/97577/elife-97577-fig6-data2-v1.zip
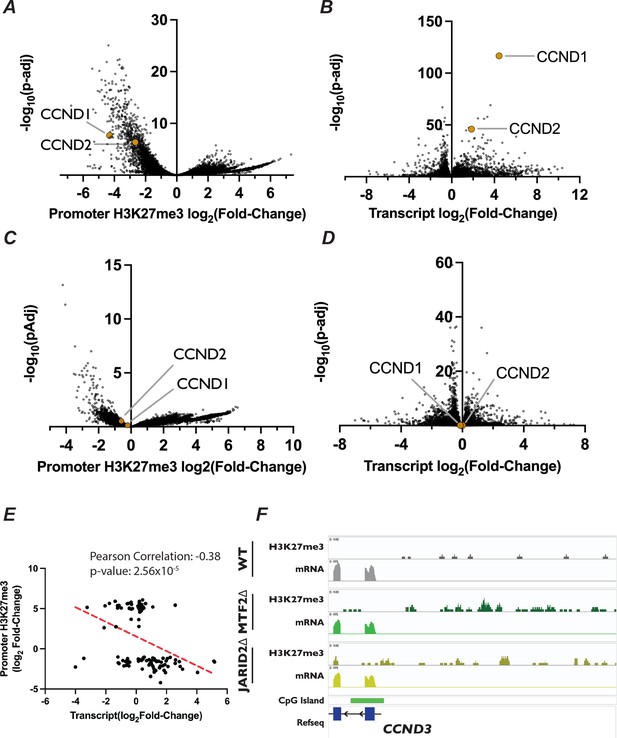
Analysis of differential H3K27me3 distribution and transcript expression of D-type cyclins in MTF2∆ and JARID2∆ cell lines.
(A) Volcano plot of DESeq2 calculated changes in log2 fold-change in H3K27me3 signal in promoters versus the log10(p-value) in enrichment in MTF2∆ cells determined by CUT&RUN. CCND1 and CCND2 location within the dataset are indicated by yellow dots. (B) Same as in (A) but for transcript abundance determined by RNA-Seq of MTF2∆ cells. (C) Volcano plot of DESeq2 calculated changes in log2 fold-change in H3K27me3 signal in promoters versus the log10(p-value) in enrichment in JARID2∆ cells determined by CUT&RUN. CCND1 and CCND2 location within the dataset are indicated by yellow dots. (D) Same as in Figure 6A but for transcript abundance determined by RNA-Seq of JARID2∆ cells. (E) Scatter plot of log2 fold-change in transcript abundance versus H3K27me3 promoter signal for genes with an adjusted p-value <0.1 in our CUT&RUN and adjusted p-value <0.05 in our RNA-Seq from JARID2∆ cell lines. (F) Genome browser traces of H3K27me3, transcript coverage, and CGI location within the CCND3 promoter region.
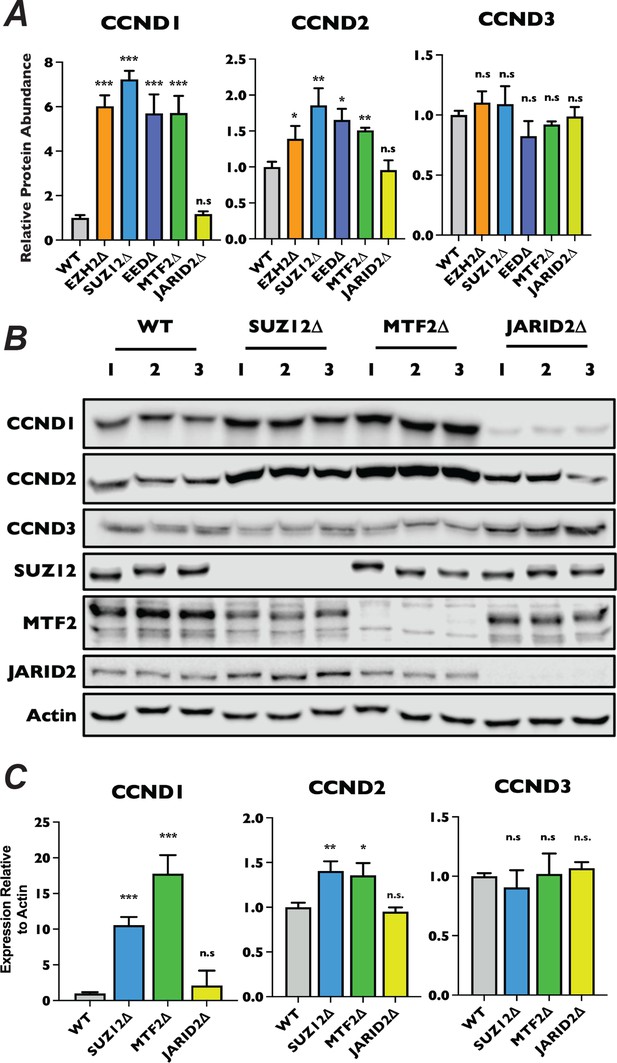
Regulation of D-type cyclin expression by MTF2-containing PRC2.1 versus PRC2.2.
(A) Quantification of protein signal from western blot in Figure 6C for CCND1 (left), CCND2 (center), and CCND3 (right) normalized to Actin. Each bar is the mean for three biological replicates, error bars ±StDev. *p-value <0.05, **p-value <0.005, ***p-value <0.0005, n.s.: not significant, two-tailed unpaired Student’s t-test. (B) Western blots of whole-cell lysates of three-independently isolated monoclonal SUZ12∆, MTF2∆, and JARID2∆ knockout cell lines probed with the indicated antibodies. (C) qRT-PCR relative quantification of CCND1, CCND2, and CCND3 mRNA levels in wild-type, SUZ12∆, MTF2∆, and JARID2∆ cells, three biological replicates, performed in technical triplicate,±StDev. *p-value <0.05, **p-value <0.005, ***p-value <0.0005, n.s.: not significant, two-tailed unpaired Student’s t-test.
-
Figure 6—figure supplement 2—source data 1
Original files for western blots shown in Figure 6—figure supplement 2B.
- https://cdn.elifesciences.org/articles/97577/elife-97577-fig6-figsupp2-data1-v1.zip
-
Figure 6—figure supplement 2—source data 2
Original files for western blots shown in Figure 6—figure supplement 2B, indicating relevant band.
- https://cdn.elifesciences.org/articles/97577/elife-97577-fig6-figsupp2-data2-v1.zip
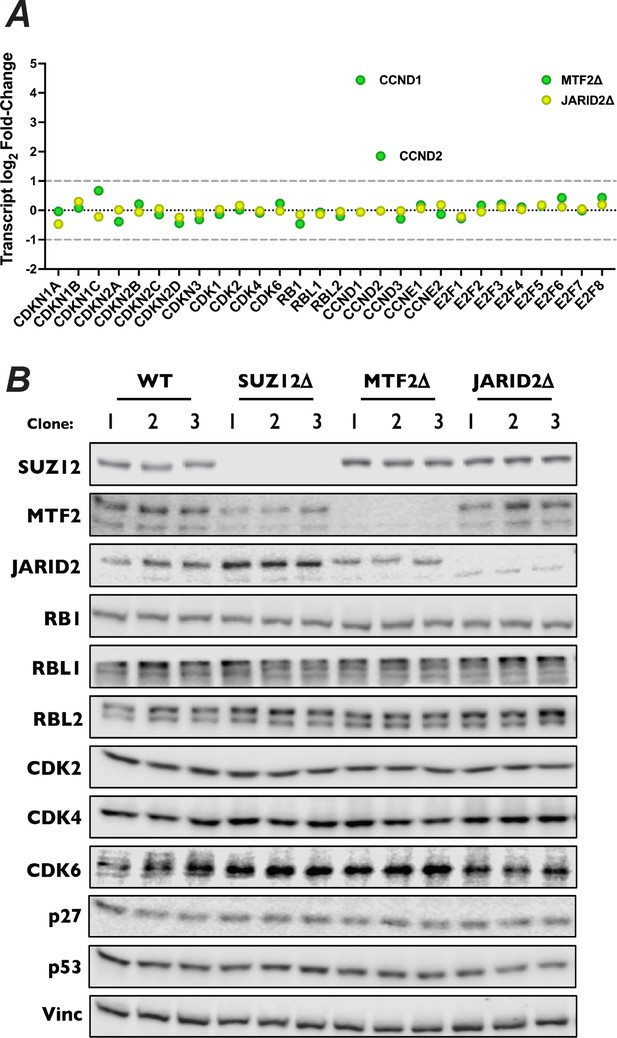
CCND1 and CCND2 display increased expression in MTF2∆ cells.
(A) Dot plot of log2 fold-change for indicated mRNAs in MTF2∆ and JARID2∆ cells. Established cut-off for significant log2 fold-change indicated by dashed gray line. (B) Western blot for a panel of G1 regulators from lysates of wild-type, SUZ12∆, MTF2∆, and JARID2∆ cell lines from three knockout cell lines probed with indicated antibodies.
-
Figure 6—figure supplement 3—source data 1
Original files for western blots shown in Figure 6—figure supplement 3B.
- https://cdn.elifesciences.org/articles/97577/elife-97577-fig6-figsupp3-data1-v1.zip
-
Figure 6—figure supplement 3—source data 2
Original files for western blots shown in Figure 6—figure supplement 3B indicating relevant band.
- https://cdn.elifesciences.org/articles/97577/elife-97577-fig6-figsupp3-data2-v1.zip
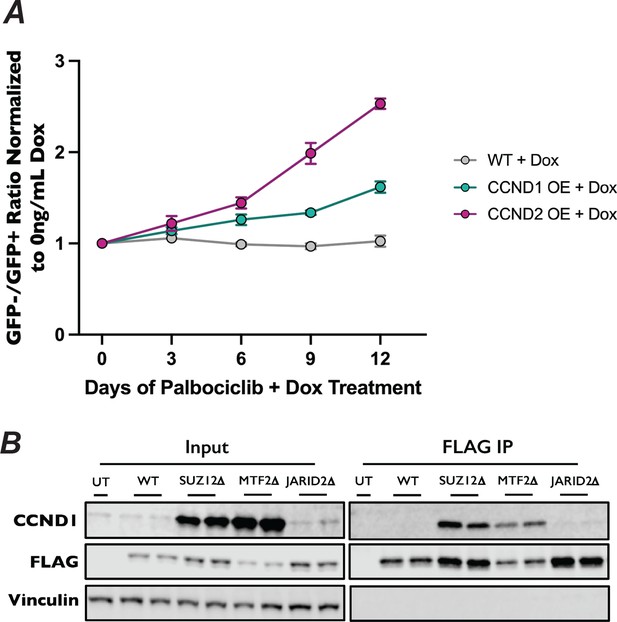
Increased expression of CCND1 and CCND2 results in resistance to palbociclib.
(A) Competitive proliferation assay of CCND1 and CCND2 overexpression cell lines resistance to palbociclib. Wild-type, dox-inducible CCND1, and dox-inducible CCND2 polyclonal HAP1 cell lines (GFP−) were mixed with wild-type cells expressing GFP (GFP+) and treated with either DMSO (mock) or 1.5 µM palbociclib, in the presence or absence of 500 ng/ml doxycycline. Mock and palbociclib-containing media, with or without doxycycline was replaced daily. Cells were split and GFP−/GFP+ ratio was assessed by flow cytometry every 3 days. Fitness of each overexpression of each pool was determined by first normalizing the GFP−/GFP+ ratio to the minus doxycycline control and then the ratio of GFP−/GFP+ between the mock and palbociclib conditions. (B) Co-immunoprecipitation of 2xFLAG-2xStrep-CDK6 expressed in wild-type, SUZ12∆, MTF2∆, and JARID2∆ knockout cell lines, probed with the indicated antibodies.
-
Figure 6—figure supplement 4—source data 1
Original files for western blots shown in Figure 6—figure supplement 4B.
- https://cdn.elifesciences.org/articles/97577/elife-97577-fig6-figsupp4-data1-v1.zip
-
Figure 6—figure supplement 4—source data 2
Original files for western blots shown in Figure 6—figure supplement 4B, indicating relevant band.
- https://cdn.elifesciences.org/articles/97577/elife-97577-fig6-figsupp4-data2-v1.zip
Additional files
-
Supplementary file 1
Orobas analysis of palbociclib, camptothecin, and colchicine chemogenetic screens.
- https://cdn.elifesciences.org/articles/97577/elife-97577-supp1-v1.xlsx
-
Supplementary file 2
Genes identified as significantly enriched or de-enriched in palbociclib, camptothecin, and colchicine chemogenetic screens.
- https://cdn.elifesciences.org/articles/97577/elife-97577-supp2-v1.xlsx
-
Supplementary file 3
DESeq2 results of promoter with significant H3K27me3 signal enrichment in CUT&RUN experiment.
- https://cdn.elifesciences.org/articles/97577/elife-97577-supp3-v1.xlsx
-
Supplementary file 4
DESeq2 results of transcripts with significant changes in RNA-Seq experiment.
- https://cdn.elifesciences.org/articles/97577/elife-97577-supp4-v1.xlsx
-
Supplementary file 5
List of cell lines, reagents, and oligonucleotides used in this study.
- https://cdn.elifesciences.org/articles/97577/elife-97577-supp5-v1.xlsx
-
MDAR checklist
- https://cdn.elifesciences.org/articles/97577/elife-97577-mdarchecklist1-v1.docx
-
Source code 1
Orobas analysis pipeline source code.
- https://cdn.elifesciences.org/articles/97577/elife-97577-code1-v1.zip





