Persistent cross-species transmission systems dominate Shiga toxin-producing Escherichia coli O157:H7 epidemiology in a high incidence region: A genomic epidemiology study
Figures
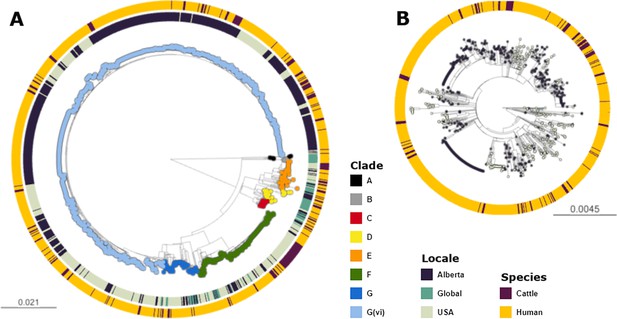
Maximum likelihood core SNP tree of the 1215 E. coli O157:H7 isolates referenced in the study.
This includes 659 isolates from Alberta, Canada, from 2007 through 2019, 494 isolates from the U.S. from 1996 through 2019, and 62 isolates from elsewhere around the globe from 2007–2016. The tree was rooted at clade A. (A) Shows all clades, with tips colored according to clade, geographic origin shown on the inner ring, and species of origin on the outer ring. (B) shows clade G(vi), which constituted 73.6% of all isolates and 88.3% of Alberta isolates. Tips are colored by geographic origin, and the ring indicates species of origin.
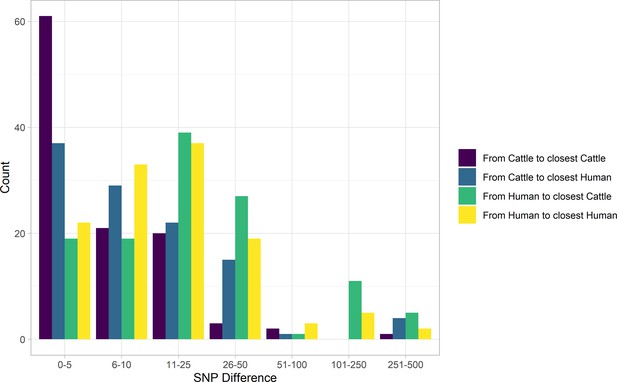
SNP distance between E. coli O157:H7 strains and their nearest relative, by species, Alberta, Canada, 2007–2015.
Distances for isolates from 121 reported human cases and 108 beef cattle are shown. Cattle isolates were highly related with 56.5% of cattle isolates within five SNPs of another cattle isolate and 94.4% within 25 SNPs. Human isolates showed a bimodal distribution in their relationship to cattle isolates, with 86.0% within 50 SNPs of a cattle isolate and the remainder 185–396 SNPs apart. Nineteen human isolates (15.7%) were within five SNPs of a cattle isolate.
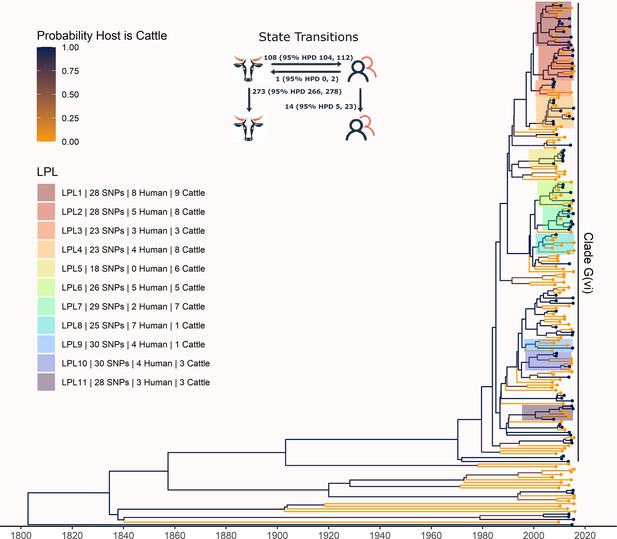
Maximum clade credibility (MCC) tree of structured coalescent analysis of E. coli O157:H7 strains isolated from 115 reported human cases and 84 beef cattle in Alberta, Canada, 2007–2015.
Isolates were down-sampled prior to phylodynamic analysis to remove isolates that were highly similar. The structured coalescent analysis estimated migration and state transitions between humans and cattle. The MCC tree was colored by inferred host, cattle (blue) or human (orange). The minimum SNP distance between all isolates in the LPL, and the number of human and cattle isolates after down-sampling are shown for each LPL. The majority of ancestral nodes inferred as cattle suggest cattle as the primary reservoir. The root was estimated at 1802 (95% HPD 1731, 1861). Eleven locally persistent lineages (LPLs) were identified, all in the G(vi) clade and labeled LPL 1 through 11. For each LPL, the minimum number of SNPs all isolates are within (30 was set as the maximum) and the number of human and cattle isolates within the LPL, not including down-sampled isolates, are shown. With down-sampled isolates reincorporated, LPLs accounted for 46 human (38.0%) and 71 cattle (65.7%) isolates. The structured coalescent model estimated 108 cattle-to-human state transitions between branches, compared to only 14 human-to-human transitions, inferring cattle as the origin of 88.5% of human lineages.
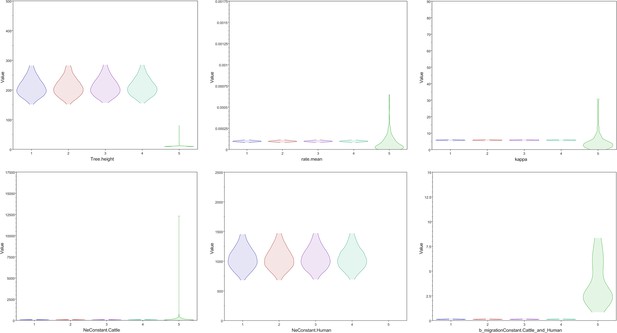
Key parameters are drawn from the posterior distributions of four Markov chain Monte Carlo chains using different starting seeds compared to draws from the prior distribution.
The posterior distributions of the four chains for the tree height, clock rate, kappa, cattle effective population size, human effective population size, and backward migration rate all differed substantially from the prior distribution, shown on the far right in green for each graph.
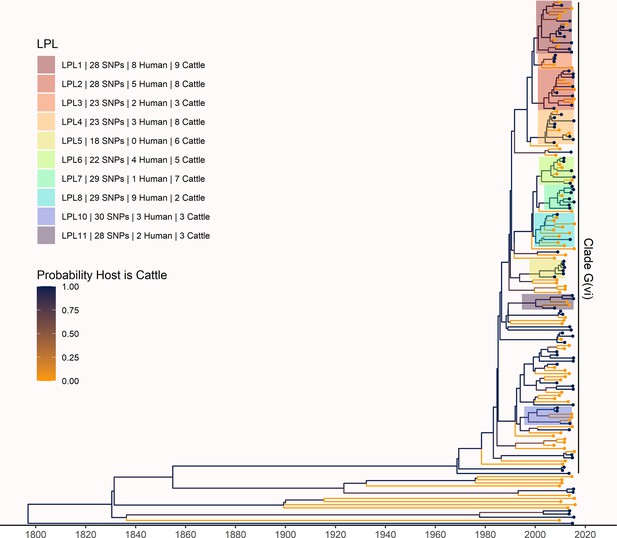
Maximum clade credibility (MCC) tree of structured coalescent analysis of 168 subsampled isolates from humans and cattle from Alberta, 2007–2015.
LPLs are labeled as in the primary analysis (Figure 3). From an initial 121 human and 108 cattle isolates, our primary analysis contained 115 human and 84 cattle isolates after down-sampling. In this sensitivity analysis, we repeated our primary analysis with a subsample of 84 randomly selected isolates from humans and the 84 cattle isolates remaining after down-sampling. As in the primary analysis, cattle were inferred as the host of the majority of ancestral nodes. The root was estimated at 1796 (95% HPD 1722, 1859), very close to the root at 1802 estimated in the primary analysis. The most recent common ancestor (MRCA) of clade G(vi) strains in Alberta was inferred to be a cattle strain, dated to 1968 (95% HPD 1956, 1979), compared to 1969 in the primary analysis. We estimated 82 (95% HPD 79, 84) human lineages arose from cattle lineages, and 5 (95% HPD 0, 11) arose from other human lineages, meaning we inferred that 94.3% of human lineages arose from cattle lineages, compared to 88.5% in the primary analysis. The LPLs identified in this sensitivity analysis were mostly identical to those identified in the primary analysis. Differences in the sensitivity analysis were that G(vi)-AB LPL 8 expanded to a larger set of isolates; G(vi)-AB LPL 9, which included only five isolates in the primary analysis, was no longer identified as an LPL, as it no longer met the five-isolate criterion; and there were minor topological changes.
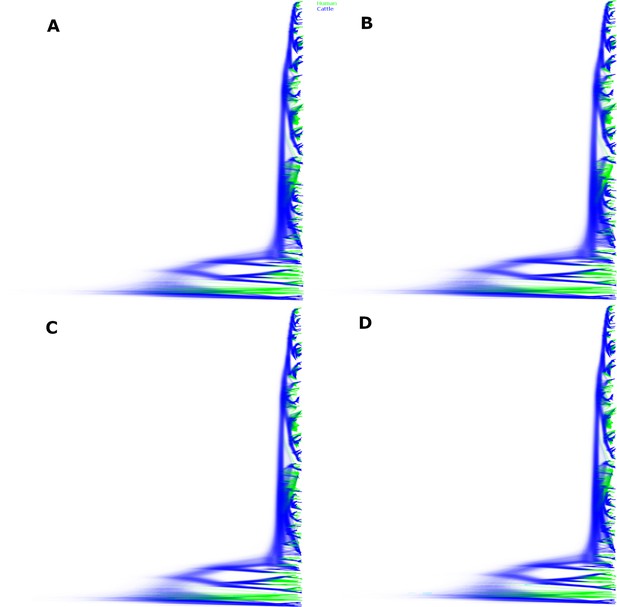
Sampled trees from four independent chains of the Alberta 2007–2015 structured coalescent analysis.
Across the four chains, 1,070,460,000 trees were sampled. Depicted here are 963,414,000 post-burn-in trees, with panels A-D each showing the trees from one chain. Lineages inferred with cattle ancestry are shown in blue, lineages inferred with human ancestry are shown in green. Reflecting the strong support for the lineages identified as locally persistent lineages (LPLs), the tree topology is well resolved.
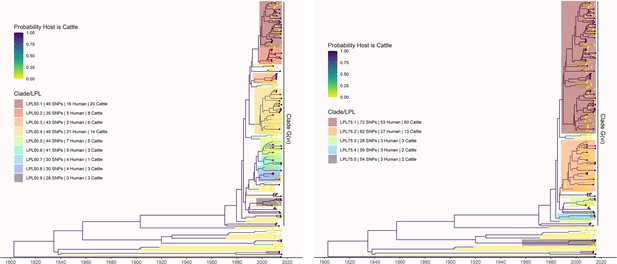
LPLs from the primary analysis were defined using alternate SNP thresholds of 50 (left) and 75 (right) SNPs.
LPL 50.1 includes G(vi)-AB LPLs 1, 2, and 3 from the primary analysis using a SNP threshold of 30. LPLs 50.2 and 50.3 correspond to G(vi)-AB LPLs 4 and 5, respectively. LPL 50.4 includes G(vi)-AB LPLs 6, 7, and 8. LPL 75.1 includes all of the above locally persistent lineages (LPLs) at the lower thresholds. All 50- and 75- threshold LPLs in this section of the tree include isolates not included at the lower SNP threshold(s). No part of LPLs 50.5 or 50.6 was identified as an LPL using a threshold of 30, but LPLs 50.7 and 50.8 correspond exactly to G(vi)-AB LPLs 9 and 10. LPL 75.2 includes LPLs 50.5–50.8 and G(vi)-AB LPLs 9 and 10. G(vi)-AB LPL 11, LPL 50.9, and LPL 75.3 are all identical. At the 75-SNP threshold, two additional LPLs are identified with no corresponding LPLs at the 30- or 50-SNP thresholds. One of these is outside the G(vi) clade.
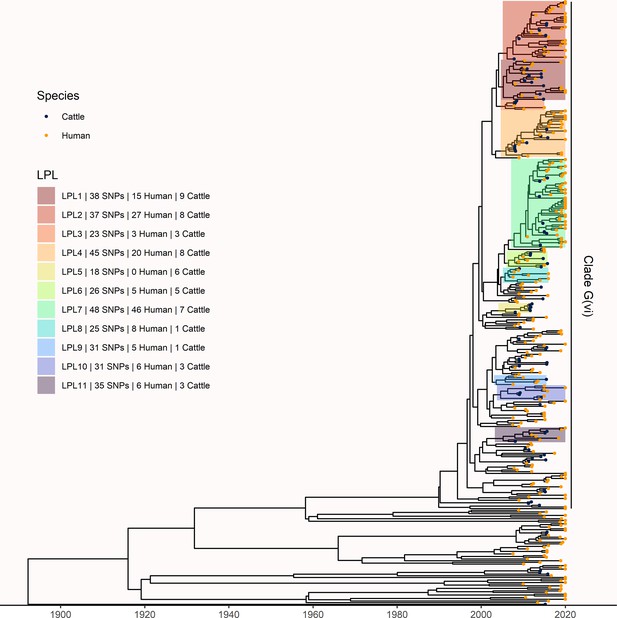
Extension of Alberta, Canada E. coli O157:H7 analysis to include 229 randomly selected study isolates and 430 additional public health isolates available from 2009–2019.
The maximum clade credibility (MCC) tree was constructed from a coalescent analysis with constant population size after down-sampling. Six locally persistent lineages (LPLs) in clade G(vi) continued to be associated with the disease after the initial study period. LPLs are colored and labeled as in Figure 3. After re-incorporating the down-sampled isolates, 74.7% of reported cases in 2018 and 2019 were associated with an LPL.
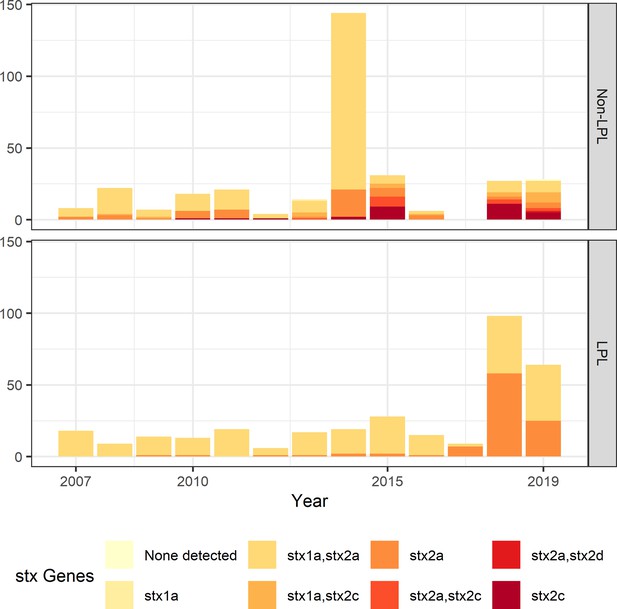
Shiga toxin gene (stx) profile by locally persistent lineage (LPL) status of extended analysis of Alberta, Canada E. coli O157:H7 isolated from cattle and humans, 2007–2019.
The stx profile across all clades shifted from the initial study period (2007–2015) to the later study period (2016–2019), with more of the virulent stx2a-only profile observed in 2018 and 2019 than in previous years. In 2018 and 2019, 51.2% of LPL isolates carried only stx2a, compared to 10.9% of non-LPL isolates. The peak in sequences in 2014 is due to two outbreaks; routine sequencing began in 2018 and 2019, accounting for the rise in sequenced cases during those years.
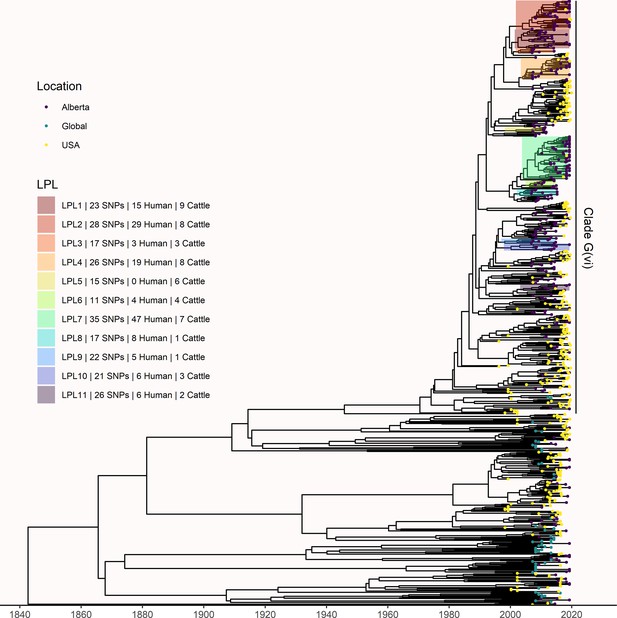
Comparison of Alberta, U.S., and global E. coli O157:H7 isolates.
Tips are colored based on isolate origin, and locally persistent lineages (LPLs) from Figure 3 are highlighted. A total of nine U.S. isolates arose from Alberta LPLs 2, 4, 7, and 11, all of which had Alberta isolates predating the U.S. isolates. No global isolates were associated with Alberta LPLs. Clade A was excluded from the analysis due to its high level of divergence from the rest of the E. coli O157:H7 population.
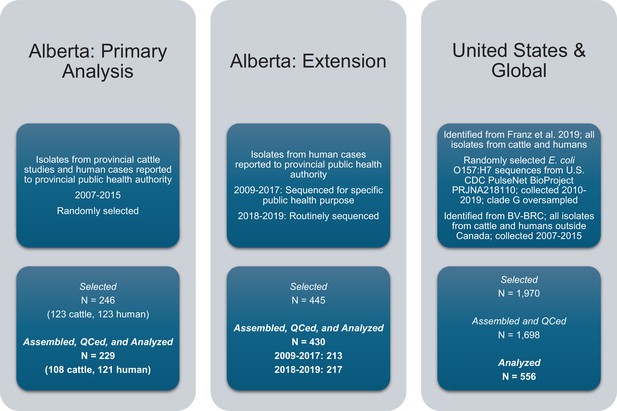
E. coli O157:H7 isolates were selected for the study and assembled.
Three sets of isolates, all originating from cattle or humans, were included in the study.
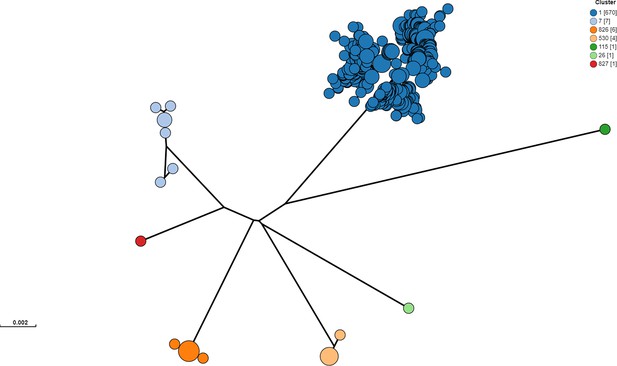
Clustering was performed from raw reads using PopPUNK v2.5.0 with 10,146 E.coli reference genomes Lees et al., 2019.
From the 246 isolates selected for sequencing for the study and 445 additional Alberta Health isolates included for contextualization, one isolate was removed prior to clustering analysis, because it was identified through metadata review as an environmental (non-human, non-cattle) isolate. Cluster 1 included the Sakai and EDL933 reference strains. Clusters 826 and 827 were novel clusters. Isolates outside of Cluster 1 were excluded from all subsequent analyses.
Tables
Distribution of study isolates by geographic source, clade, and Shiga toxin gene (stx) profile.
| Clade | Source | stx1a | stx1a/stx2a | stx1a/stx2c | stx1a/stx2a/stx2c | stx2a | stx2c | stx2a/stx2c | stx2a/stx2c/stx2d | stx2a/stx2d | None detected | Total |
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| G(vi) | 2 | 682 | 0 | 0 | 210 | 0 | 0 | 0 | 0 | 0 | 894 | |
| Alberta | 0 | 443 | 0 | 0 | 139 | 0 | 0 | 0 | 0 | 0 | 582 | |
| The U.S. | 2 | 237 | 0 | 0 | 71 | 0 | 0 | 0 | 0 | 0 | 310 | |
| Global | 0 | 2 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 2 | |
| Other G | 3 | 1 | 4 | 2 | 4 | 33 | 6 | 0 | 0 | 5 | 58 | |
| Alberta The | 0 | 0 | 4 | 0 | 0 | 10 | 1 | 0 | 0 | 0 | 15 | |
| U.S. | 2 | 1 | 0 | 2 | 1 | 19 | 4 | 0 | 0 | 3 | 32 | |
| Global | 1 | 0 | 0 | 0 | 3 | 4 | 1 | 0 | 0 | 2 | 11 | |
| F | 0 | 0 | 0 | 0 | 53 | 23 | 75 | 5 | 1 | 7 | 164 | |
| Alberta The | 0 | 0 | 0 | 0 | 12 | 9 | 12 | 0 | 1 | 1 | 35 | |
| U.S. | 0 | 0 | 0 | 0 | 33 | 11 | 61 | 5 | 0 | 6 | 116 | |
| Global | 0 | 0 | 0 | 0 | 8 | 3 | 2 | 0 | 0 | 0 | 13 | |
| Other (A-E) | 2 | 1 | 41 | 0 | 1 | 50 | 2 | 0 | 0 | 2 | 99 | |
| Alberta The | 1 | 0 | 15 | 0 | 0 | 11 | 0 | 0 | 0 | 0 | 27 | |
| U.S. | 0 | 0 | 10 | 0 | 1 | 24 | 1 | 0 | 0 | 0 | 36 | |
| Global | 1 | 1 | 16 | 0 | 0 | 15 | 1 | 0 | 0 | 2 | 36 | |
| Total | 7 | 684 | 45 | 2 | 268 | 106 | 83 | 5 | 1 | 14 | 1215 |
Estimated migrations from structured coalescent analysis of Alberta, U.S., and global isolates, excluding clade A.
| Migration direction | Mean | 95% HPD |
|---|---|---|
| Alberta to Alberta | 589 | 570, 609 |
| Alberta to Global | 8 | 4, 11 |
| Alberta to U.S. | 12 | 6, 16 |
| Global to Alberta | 68 | 57, 78 |
| Global to Global | 442 | 363, 511 |
| Global to U.S. | 296 | 267, 335 |
| U.S. to Alberta | 5 | 0, 14 |
| U.S. to Global | 13 | 0, 35 |
| U.S. to U.S. | 104 | 10, 184 |
-
HPD, highest posterior density interval.
Analyses conducted and model priors.
| Analysis | Isolates included | Isolates remaining after down-sampling | Tree model | Substitution model | Clock model |
|---|---|---|---|---|---|
| Primary analysis* | Alberta 2007–2015 (n=229) | 115 human 84 cattle | Structured coalescent with two demes (1 per species); Ne initial value 10 and Weibull distribution (shape = 1, scale = 100); migration initial value 1.0 and Exponential distribution (mean = 1) | HKY with empirical frequencies and discrete Gamma site model with four categories | Relaxed log-normal with initial value 1.5×10–5 and Log-Normal distribution (M=1.5×10–5, S=1.5, with mean in real space) |
| Alberta long-term persistence† | Alberta 2007–2019 (n=657) | 274 human 84 cattle | Coalescent constant population with Ne initial value 10 and Weibull distribution (shape = 1, scale = 100) | HKY with empirical frequencies and discrete Gamma site model with four categories | Relaxed log-normal with initial value 1.5×10–4 and Log-Normal distribution (M=1.5×10–4, S=1.5, with mean in real space) |
| Global circulation (unstructured)† | Alberta 2007–2019 (n=657) The U.S. 1999–2019 (n=492) Global 2007–2016 (n=61) | Alberta: 274 humans, 84 cattle U.S.: 312 humans, 38 cattle Global: 39 humans, 22 cattle | Coalescent constant population with Ne initial value 10 and Weibull distribution (shape = 1, scale = 100) | HKY with empirical frequencies and discrete Gamma site model with four categories | Relaxed log-normal with initial value 1.5×10–4 and Log-Normal distribution (M=1.5×10–4, S=1.5, with mean in real space) |
| Global circulation (structured)† | Alberta 2007–2019 (n=657) The U.S. 1999–2019 (n=492) Global 2007–2016 (n=61) | Alberta: 274 humans, 84 cattle U.S.: 312 humans, 38 cattle Global: 39 humans, 22 cattle | Structured coalescent with three demes (1 per geography); Ne initial value 10 and Weibull distribution (shape = 1, scale = 100); migration initial value 1.0 and Exponential distribution (mean = 1) | HKY with empirical frequencies and discrete Gamma site model with four categories | Relaxed log-normal with initial value 1.5×10–4 and Log-Normal distribution (M=1.5×10–4, S=1.5, with mean in real space) |
-
*
A primary analysis was run using four different random seeds to confirm that all converged to the same solution. The four runs were combined to produce the final maximum clade credibility tree and state transition estimates. Model priors from this analysis were also used in an analysis in which draws were taken from the prior distribution, as opposed to the posterior distribution, to confirm that the final results were not overly influenced by the choice of priors.
-
†
Clade A isolates were excluded from these analyses given the very small number available from any locale and clade A’s divergence from the rest of the clades.
Additional files
-
MDAR checklist
- https://cdn.elifesciences.org/articles/97643/elife-97643-mdarchecklist1-v1.pdf
-
Source data 1
List of accession numbers for sequences used with deidentified metadata.
- https://cdn.elifesciences.org/articles/97643/elife-97643-data1-v1.xlsx





