Light-driven synchronization of optogenetic clocks
Figures
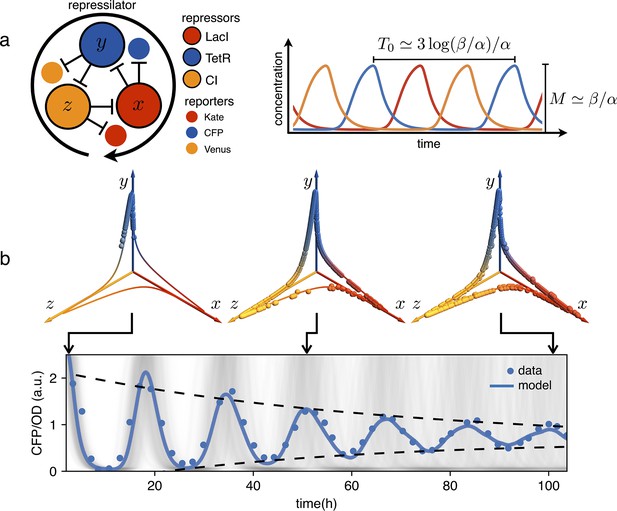
Dephasing in a population of repressilators.
(a) (left) Schematic representation of the repressilator 2.0 plasmid. (right) Simulated time evolution of the three repressors concentrations in the limit cycle of the protein-only model in Equation 1. The reported expressions for amplitude and period are derived in the limits and (see Appendix 1-section 1). (b) Concentration of the reporter CFP (TetR) as a function of time for an initially synchronized population of oscillators growing in a multi-well plate. Blue circles represent average over two technical replicates, corresponding errorbars are comparable to symbol size. The shadowed gray curves show simulations of independent oscillators (see Materials and methods) and standard deviation 0.034 h-1. Dephasing due to the variability of oscillators periods causes a damping in the mean population signal reported as a blue line. Snapshots of the population ensemble in the 3D concentration space are reported for three instants of time during the simulation, clearly showing progressive dephasing of individual oscillators over the limit cycle trajectory.
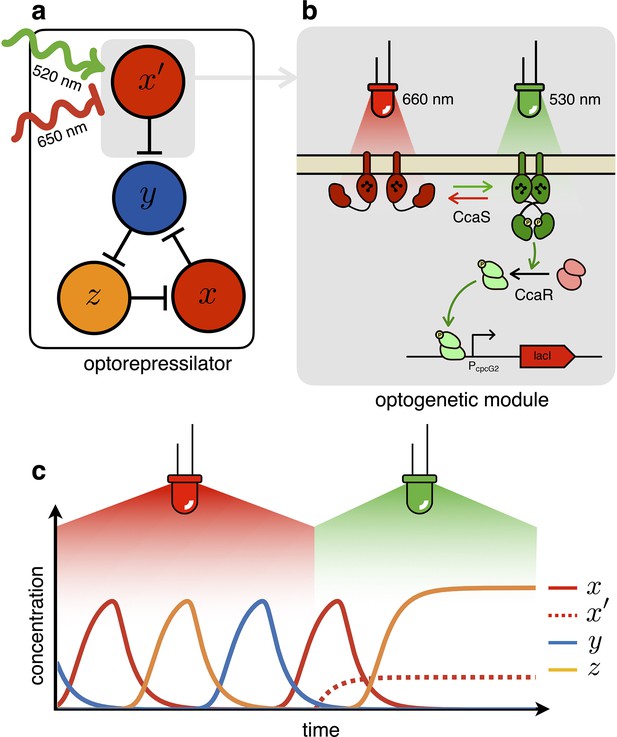
The optorepressilator: a light-controllable genetic oscillator.
(a) Schematic illustration of the optorepressilator circuit. Green and red light respectively promote and repress the production of one of the three transcription factors in the repressilator (LacI). (b) Working mechanism of the optogenetic module. A light-driven two-component system controls the production of an additional copy of lacI. The optogenetic system consists of the membrane-associated histidine kinase CcaS and its response regulator CcaR. Absorption of green light increases the rate of CcaS autophosphorylation and phosphate transfer to the transcription factor CcaR. Phosphorylated CcaR promotes transcription of an additional copy of the LacI gene from the promoter . Red light reverts CcaS to the inactive state and shuts down transcription from . (c) Ideal optorepressilator’s dynamics according to Equation 2. The system oscillates unperturbed under red light, but collapses to a fixed point under green light when the extra represses .
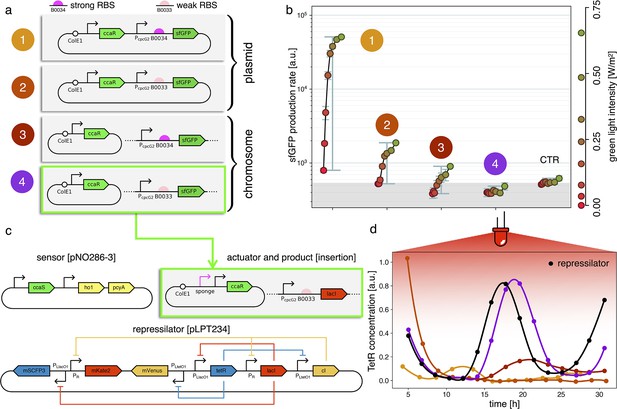
Fine-tuning gene expression in the optorepressilator circuit.
(a) To tune expression from the optogenetic module we varied the strength of RBSs and gene copy number by placing the output gene (sfGFP) either on a plasmid or in the chromosome. (b) Light-controlled expression levels of the sfGFP reporter from the four constructs in (a) and a negative control consisting of DHL708 with plasmids pNO286-3 and pSR58-0 (optogenetic plasmids without sfGFP cassette). For each curve, the samples were exposed to a fixed level of red light (intensity 0.74 W/m2) while increasing green light intensity, as indicated by the dots’ colors (color reference on the right). The main error bars represent the dynamic range of each sample. The smaller error bars represent the standard deviation of individual light conditions between replicates made in three separate days. The gray shaded area represents the autofluorescence background. Light intensities and raw fluorescence data are in Appendix 1—figure 6. (c) Scheme of the final optorepressilator circuit (see Materials and methods for details). (d) Time evolution of TetR reporter (CFP) concentration in a population of IPTG synchronized cells growing under red light. Black points from the original repressilator 2.0 display marked oscillations. Colored lines correspond to the four constructs in a with sfGFP replaced by LacI and the addition of the sponge promoters as in pSpongeROG. Circles represent data, where each dot is the average of two replicates with error bars comparable to marker size, while lines are spline interpolations. Only the purple line, corresponding to the system in (c), shows oscillations comparable to those of the original repressilator. The difference in period can be explained by the difference in growth rate of the two strains, as shown in Appendix 1—figure 4.
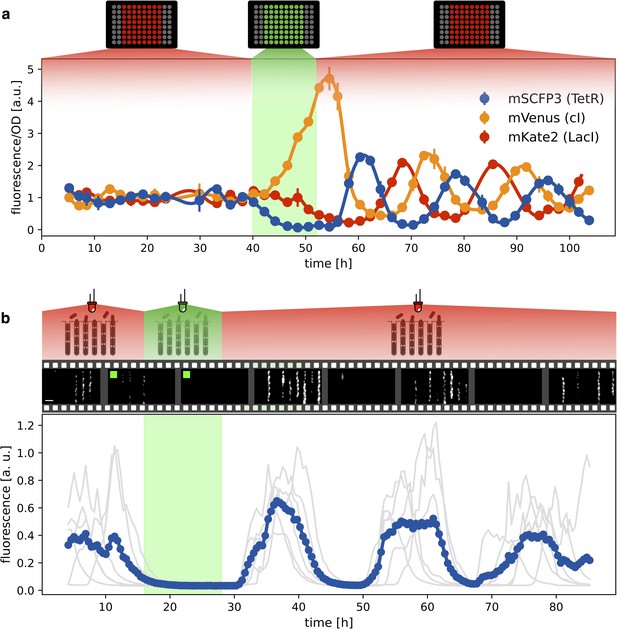
Optical synchronization.
(a) Time evolution of mSCFP3(CFP), mVenus and mKate2 signals from a population of exponentially growing cells in multiwell plate. Red and green shaded areas represent the illumination protocol. Dots reports data for optorepressilator cells and clearly shows the appearance of synchronous oscillations after transient illumination with green light, each marker is the average of two replicates and the error bar is data range. (b) Single cell data from a mother machine experiment employing a similar light protocol as in (a) (Video 1). The gray curves represent the concentration of CFP for individual cells growing in different channels, while the blue curve is their average. Snapshots above the plot report fluorescence imaging of bacteria in the microfluidic chip, centered at the corresponding time point on the time axis below. Scale bar is 5 μm.
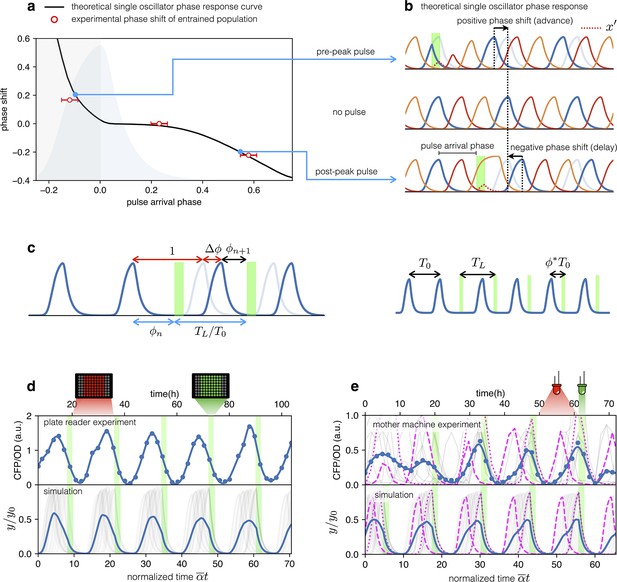
Optical entrainment.
(a) Phase shift as a function of pulse arrival phase. (b) Numerical simulations of Equation (2) illustrating the effect of pulse arrival phase on the phase of optorepressilator oscillations. (c) Graphical illustration of the parameters used to describe the phase shift. (d) (top) CFP signal from a population of cells growing exponentially in 96-well plates that are exposed to a train of periodic pulses of green light. The population signal displays undamped oscillations demonstrating optical entrainment. (bottom) Numerical simulation of an ensemble of optorepressilators evolving according to Equation (2) and with distributed growth rates (same parameters as in Figure 1b). Pulses have a duration of and a relative period of . Gray lines are individual oscillators while blue is the population average. (e) (top) Gray lines are CFP signals from single cells growing in a mother machine under periodic pulsed illumination (Video 2). The blue curve is the average. (bottom) Numerical simulation with the same parameters as in (d) but a larger dispersion of growth rates and a slightly shorter relative period was required to match experiments above. In both data and simulation the magenta dotted curve highlights a slow oscillator (low growth rate and larger amplitude) entrained with a phase such as to receive the light pulse on the rising edge to anticipate the next oscillation. Magenta dashed lines highlight faster oscillators whose phase is such to receive the pulse on the decaying edge to delay the following oscillation.
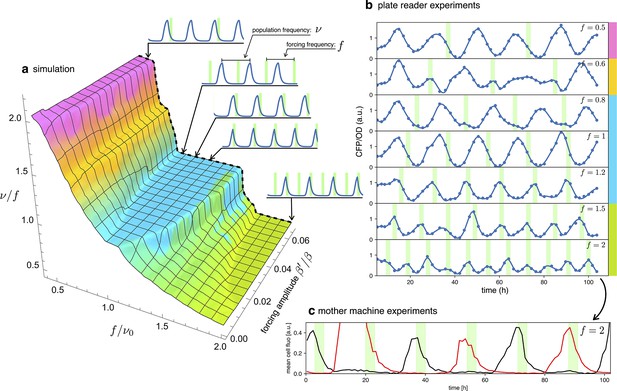
Detuning.
(a) 3D surface representing the ratio between the actual frequency of a single forced optorepressilator and the frequency of the forcing signal, as a function of the forcing frequency and amplitude. The colors of the plateaus (Arnold’s tongues) highlight different regions of global synchronization in the frequency/amplitude plane. (b) Time evolution of the average CFP concentration (blue curve) in a plate reader experiment, for different frequencies of green light pulses (green bars). The color bars at the right of the plot indicates the corresponding region on the surface in (a). (c) Mother machine experiment revealing two distinct cells oscillating at half the signal frequency but peaking on alternate pulses of incoming light. This explains why for and in (b) we observe a population mean oscillating with the same frequency of the forcing instead of the value predicted for the green tongue in (a).
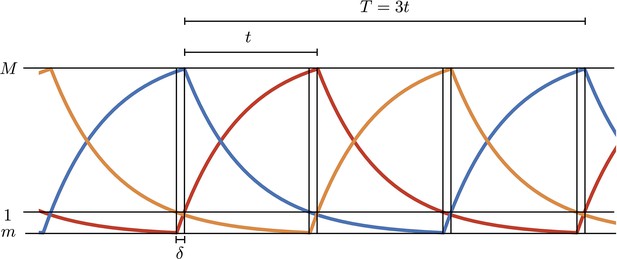
Simulations of the digital model.
and , respectively, are the maximum and minimum values in the oscillations and is the time interval to rise from the minimum to 1 (threshold value in units).
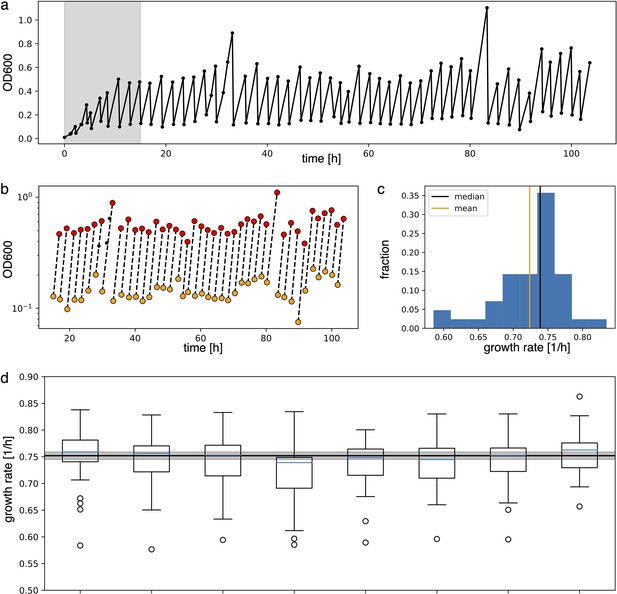
Analysis to estimate growth rate from experimental data.
(a) Raw absorbance data from one individual sample in the plate reader experiment. The initial gray area is removed from further analysis to ensure samples reached stable growth conditions. (b) Minimum and maximum OD600 points (respectively in yellow and red) in log scale are fitted with a line to extract the growth rate as its slope. (c) Distribution of the growth rates. Median and mean of the distribution are shown respectively in black and orange. Due to the non-symmetric distribution, we used the median to summarize the growth rate. (d) Box plots of the growth rates for each sample, all grown in parallel. The blue line inside the box plots is the median. The black line is the mean of all medians, our estimate growth rate () for the optorepressilator in plate reader experiments. The gray area represents the mean growth rate ± the standard deviation.
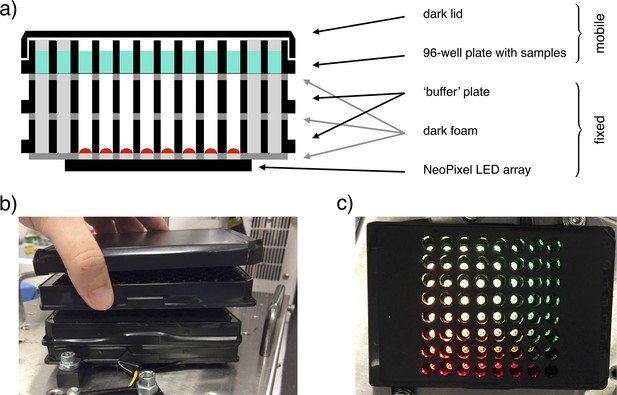
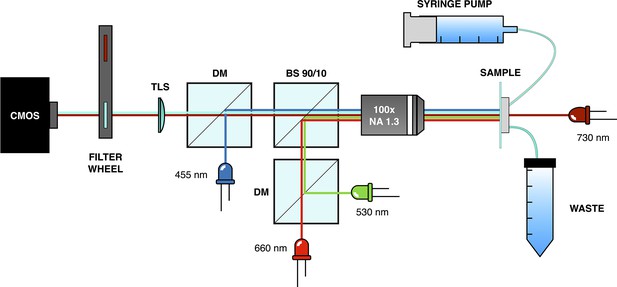
Illustration of the light-addressable multiwell plate.
(a) Scheme of a longitudinal section of the device, with key components highlighted. (b) Frontal picture of the custom-made light-addressable multiwell plate placed in a shaking incubator. The sample plate and dark lid are partially lifted. (c) Top-down picture of the device. Dark lid and sample plate are removed and LEDs are on.
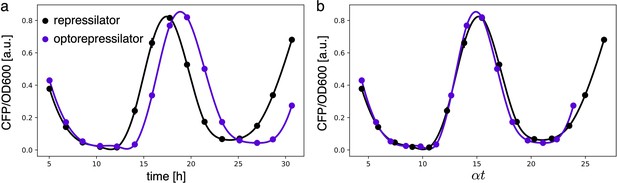
Period of different constructs overlaps when normalized by growth rate.
(a) Fluorescence intensities of the repressilator and optorepressilator, as shown in Figure 3d. (b) Time axis rescaled: time points of samples in a multiplied by their respective growth rates (0.87 h-1, 0.76 h-1) and aligned to a common initial time. Post-rescaling, the oscillations of the repressilator and optorepressilator overlap nicely.
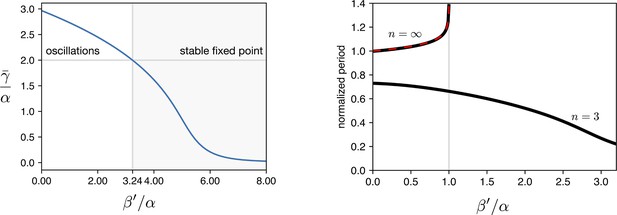
Simulations of leakage’s effect on limit cycle and period.
(a) vs leakage shows that increasing the leakage past a certain threshold the existing limit cycle breaks down and the system does damped oscillations towards the fixed point. (b) The period of the oscillator is affected by the leakage. We plot the curves for the two hill coefficients treated in the manuscript. For the period is monotonically decreasing in response to leakage increase, suggesting that the period increase in the optorepressilator respect to the repressilator (Figure 3d main text) is not leakage. In the digital case () we see that the leakage breaks the limit cycle for values of .
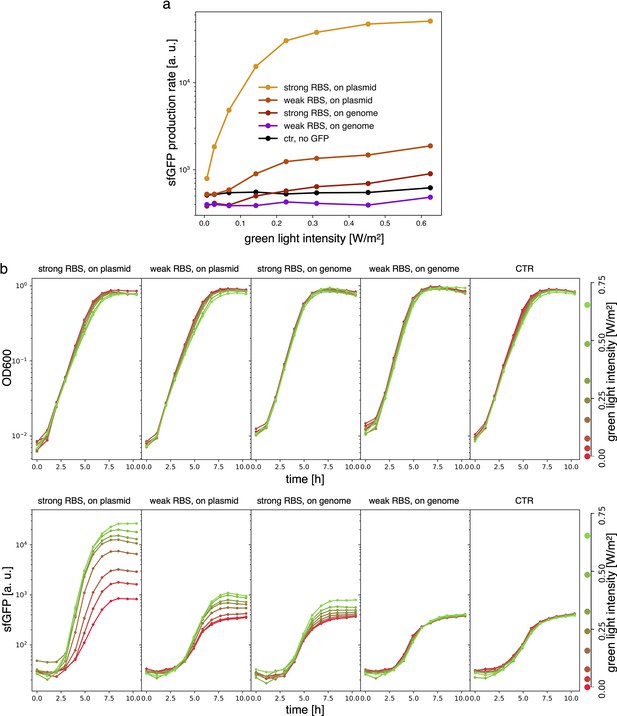
Samples are DHL708 E. coli strain with pNO286-3 plasmid, added with sfGFP light-driven production construct as shown in Figure 3 of the main text, while control (’ctr’) is DHL708+pNO286-3+pSR58-0.
(a) Protein production rate over green light intensity (same data as Figure 3b). While being exposed to different green light intensities, all samples were also constantly exposed to 0.74 W/m2 red light. Dots are the mean values of experiments repeated on 3 different days. (b) Raw OD600 (top) and sfGFP fluorescence (bottom) data for one of the sfGFP production replicates.
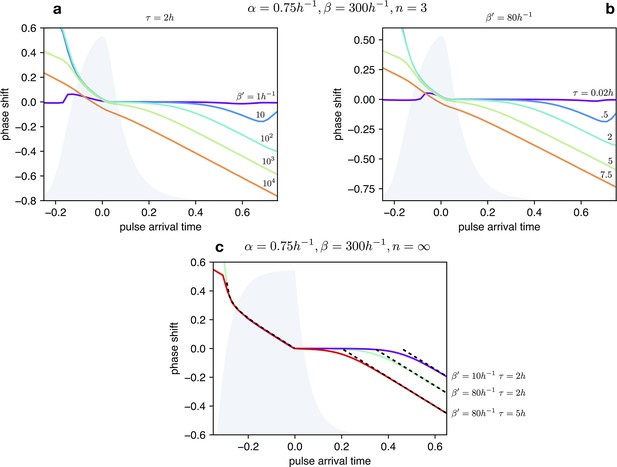
Impact of light dependent production rate β’ and input duration τ on Phase Response Curves.
(a) PRCs of the optorepressilator for h and varying logarithmically between 1 h-1 and 104 h-1. (b) PRCs of the optorepressilator for h-1 and varying between 0.02 h and 75 h. (c) PRCs of the optorepressilator in the digital approximation () for varying and . The black dashed lines represent the theoretical prediction as described below. The other parameters used in the simulations are h-1, h-1, .
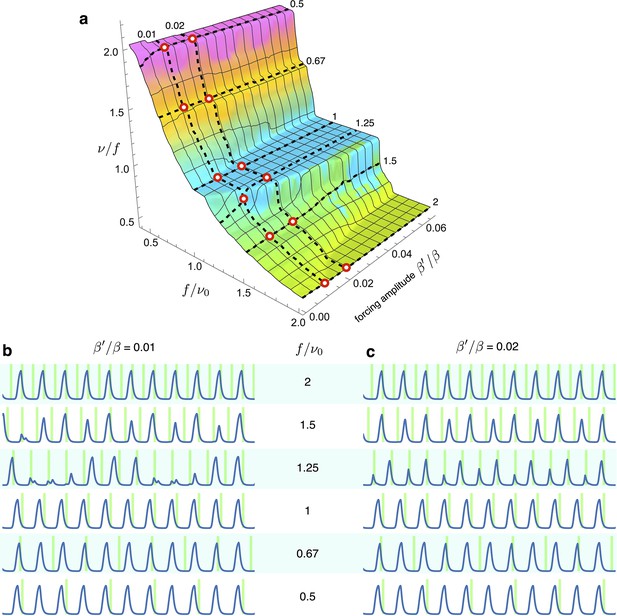
Simulations of optorepressilators’ response to low forcing amplitude at various frequencies.
(a) Arnold surface calculated as in Figure 6 of main text, with red circles to highlight the values of forcing amplitudes and frequencies used for plots in (b and c). We notice that at a forcing frequency ratios of 1.5 and 1.25, entrainment is achieved only for the higher amplitude values. Interestingly, when moving in between the main plateaus in the Arnold surface we find entrainment at fractional frequency ratios such as at and at .
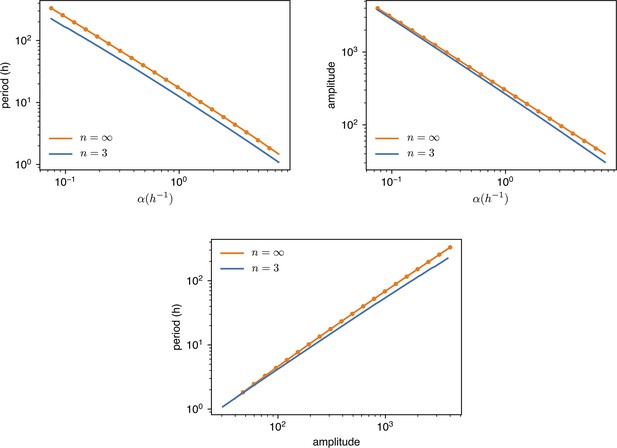
values are changed from of the standard value () to 10 times as much.
Amplitude is dimensionless, in units of . Dots are simulations, the continuous lines are predictions with or digital approximation .
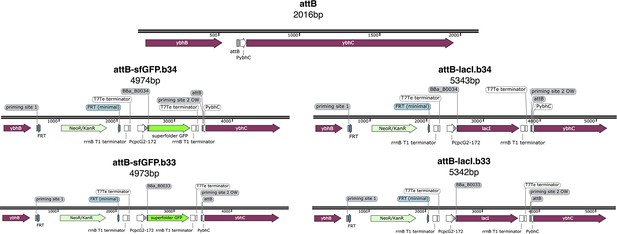
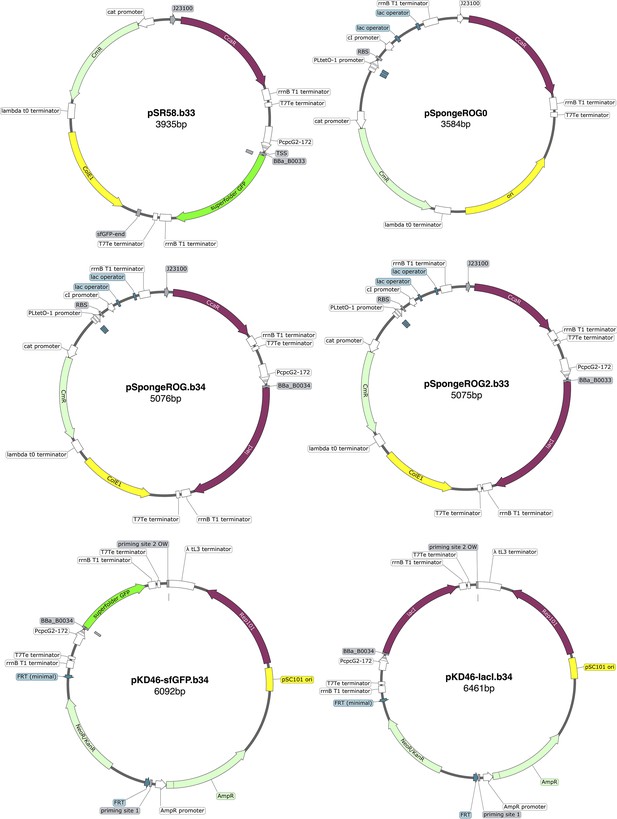
Maps of the original attB site and genome insertions.
Genome sites correspond to E. coli strains: MCC0034 top left, MCC0033 bottom left, MCC0234 top right, MCC0233 bottom right. All maps are created with SnapGene.
Videos
Optical synchronization in mother machine.
The snapshots of CFP fluorescence were acquired every 9 minutes and the video is played at 32 frames/s. The presence of a green square over the image indicates that at that time point the bacteria are exposed to green light stimuli provided by the Thorlabs LED M530L4. The fluorescence of each mother cell and the mean fluorescence of all mother cells are plotted in Figure 4b. Scale bar is 5 μm.
Optical entrainment in mother machine.
The snapshots of CFP fluorescence were acquired every 9 minutes and the video is played at 32 frames/s. The presence of a green square over the image indicates that at that time point the bacteria are exposed to green light stimuli provided by the Thorlabs LED M530L4. The fluorescence of each mother cell and the mean fluorescence of all mother cells are plotted in Figure 5e. Scale bar is 5 μm.
Tables
| Reagent type (species) or resource | Designation | Source or reference | Identifiers | Additional information |
|---|---|---|---|---|
| Strain, strain background (Escherichia coli) | DHL708 | Potvin-Trottier et al., 2016 | Addgene_98417 | |
| Recombinant DNA reagent | pLPT234 (plasmid) | Riglar et al., 2019 | RRID:Addgene_127855 | |
| Recombinant DNA reagent | pLPT145 (plasmid) | Potvin-Trottier et al., 2016 | RRID:Addgene_85527 | |
| Recombinant DNA reagent | pNO286-3 (plasmid) | Ong and Tabor, 2018 | RRID:Addgene_107746 | |
| Recombinant DNA reagent | pSR58.6 (plasmid) | Schmidl et al., 2014 | RRID:Addgene_63176 | |
| Recombinant DNA reagent | pKD46 (plasmid) | Datsenko and Wanner, 2000 | ||
| Recombinant DNA reagent | pKD4 (plasmid) | Datsenko and Wanner, 2000 | RRID: Addgene_45605 | |
| Commercial kit | In-Fusion HD Cloning | Takara Bio |
Escherichia coli strains used in this work.
| Bacterial strain | Genotype | Source | Parent |
|---|---|---|---|
| DHL708 | MC4100 | Potvin-Trottier et al., 2016 | |
| MCC0234 | DHL708 | this work | DHL708 |
| MCC0233 | DHL708 | this work | DHL708 |
| MCC0034 | DHL708 | this work | DHL708 |
| MCC0033 | DHL708 | this work | DHL708 |
Plasmids.
| Plasmid | Description | Source | Parent |
|---|---|---|---|
| pLPT234 | core repressilator componentsand reporters | Riglar et al., 2019 | |
| pLPT145 | repressors’ sponge | Potvin-Trottier et al., 2016 | |
| pNO286-3 | light sensor andPCB enzymes | Ong and Tabor, 2018 | |
| pSR58.6 | CcaR, light-drivensfGFP production(RBS BBa_B0034) | Schmidl et al., 2014 | |
| pSR58.b33 | CcaR, light-drivensfGFP production(RBS BBa_B0033) | this work | pSR58.6 |
| pSR58-0 | CcaR, no nor sfGFP | this work | pSR58.6 |
| pSpongeROG.b34 | light-driven LacI production (RBS BBa_B0034), repressors’s sponge, CcaR | this work | pSR58.6 (backbone), pLPT145 (sponge), pLPT234 (lacI) |
| pSpongeROG.b33 | light-driven LacIproduction(RBS BBa_B0033),repressors’s sponge,CcaR | this work | pSR58.6 (backbone), pLPT145 (sponge), pLPT234 (lacI) |
| pSpongeROG0 | repressors’s sponge,CcaR | this work | pSR58.6 (backbone), pLPT145 (sponge) |
| pKD46-sfGFP.b34 | genome insertionof light-drivensfGFP cassette | this work | pKD46 (backbone),pSR58.6 |
| pKD46-sfGFP.b33 | genome insertionof light-drivensfGFP cassette | this work | pKD46 (backbone),pSR58.b33 |
| pKD46-lacI.b34 | genome insertionof light-drivenlacI cassette | this work | pKD46 (backbone),pSpongeROG.b34 |
| pKD46-lacI.b33 | genome insertionof light-drivenlacI cassette | this work | pKD46 (backbone),pSpongeROG.b33 |
Antibiotic concentrations.
| Antibiotic | Working concentration |
|---|---|
| Streptomycin | 50 μg/ml |
| Ampicillin | 100 μg/ml |
| Kanamycin | 50 μg/ml |
| Chloramphenicol | 20 μg/ml |
| Spectinomycin | 100 μg/ml |
Additional files
-
MDAR checklist
- https://cdn.elifesciences.org/articles/97754/elife-97754-mdarchecklist1-v1.pdf
-
Source data 1
Plate reader data of optorepressilator detuning (Figure 6b, Figure 5d), optorepressilator light-driven synchronization (Figure 4a) and of repressilator desynchronization (Figure 1b).
- https://cdn.elifesciences.org/articles/97754/elife-97754-data1-v1.xlsx
-
Source data 2
Plate reader data of GFP protein expression levels (Figure 3b).
- https://cdn.elifesciences.org/articles/97754/elife-97754-data2-v1.xlsx
-
Source data 3
- https://cdn.elifesciences.org/articles/97754/elife-97754-data3-v1.xlsx
-
Source data 4
Plate reader data of the comparison between optorepressilator versions (Figure 3d).
- https://cdn.elifesciences.org/articles/97754/elife-97754-data4-v1.xlsx