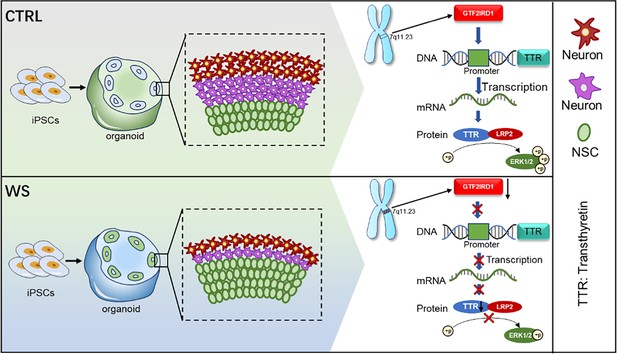
A human forebrain organoid model reveals the essential function of GTF2IRD1-TTR-ERK axis for the neurodevelopmental deficits of Williams syndrome
Figures
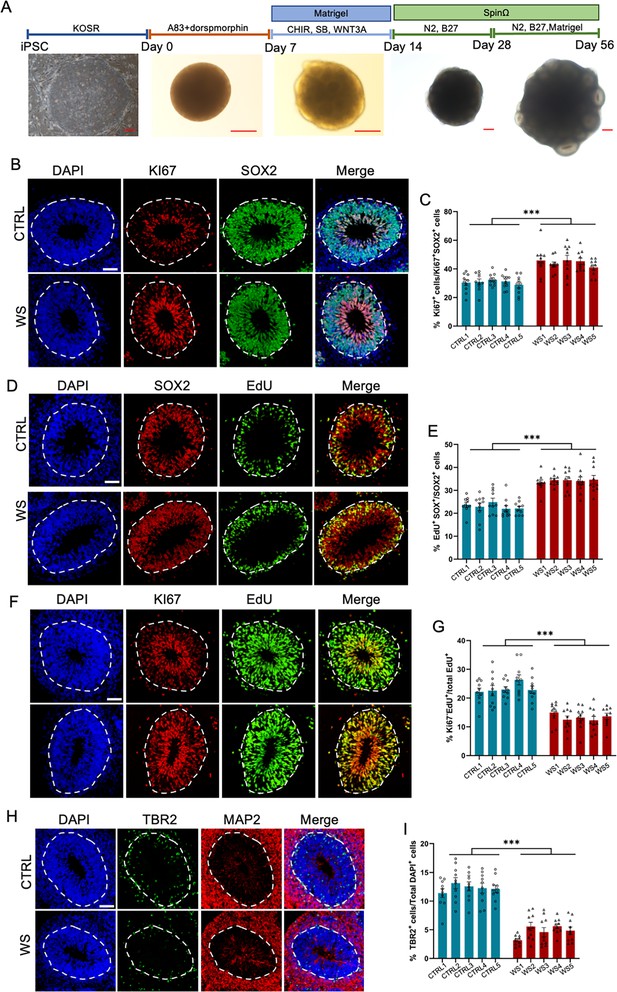
WS neural progenitor cells display abnormal proliferating capability.
(A) Schematic illustration of forebrain organoids culture process and representative brightfield images of organoids over development. Scale bar, 200 μm. (B) Representative images of KI67 and neural progenitor cells (NPCs) marker SOX2 immunofluorescence staining with healthy (CTRL) and WS patient (WS) forebrain organoids at day 56, respectively. The proliferating cells were positive for KI67 labeling, and NPCs were positive for SOX2 labeling. The white dotted line indicated the outer interface of the ventricular zone (VZ) region. (C) Quantification results showed that the average proportion of KI67 positive (KI67+) cells among total NPCs (SOX2+) significantly increased in the VZ layer of WS forebrain organoids relative to CTRL. (D) Representative images of EdU and SOX2 immunofluorescence staining with CTRL and WS forebrain organoids at day 56. EdU was applied for 2 hr. (E) Quantification results showed that the average proportion of EdU+ cells among total SOX2+ NPCs significantly increased in the VZ layer of WS forebrain organoids compared to CTRL. (F) Representative images of EdU and KI67 immunofluorescence staining with CTRL and WS forebrain organoids at day 56, respectively. EdU was applied for 24 hr. (G) Quantification results show that the average proportion of Ki67-EdU+ cells among total EdU+ cells significantly decreased in the VZ layer of WS forebrain organoids compared to CTRL. EdU was applied for 24 hr. Ki67-EdU+ cells were counted, that is, the proportion of cells exiting the cell cycle. (H) Representative images of TBR2, a marker for intermediate precursor cells (IPCs), and neuronal marker MAP2 immunofluorescence staining of CTRL and WS forebrain organoids, respectively. (I) Quantification results showed that the average proportion of TBR2+ IPCs remarkably decreased in WS forebrain organoids compared to CTRL. For (B-I), five IPS lines of three healthy control (Ctrl) and three WS patients (WS) were adopted for the study, respectively. Three to five organoids were analyzed from each IPS line and 10 representative rosettes of organoids were analyzed in each line. values represent mean ± SEM. *p<0.05, **p<0.01, ***p<0.001, one-way ANOVA followed by Dunnett’s multiple-comparison test. Scale bar, 50 μm.
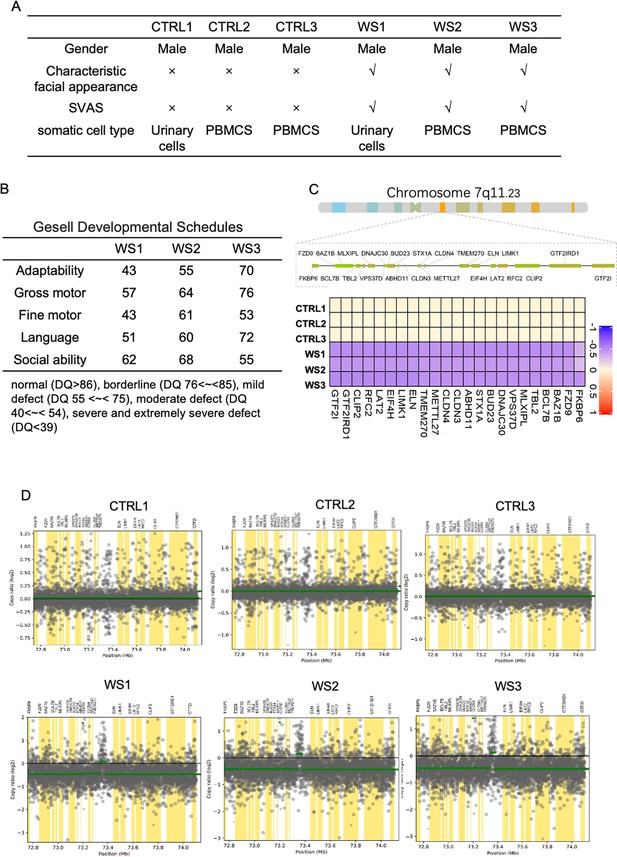
Clinical information and genetic analysis of CTRL and WS donors.
(A) Basic clinical data of three healthy controls (CTRL) and three WS patients. Characteristic facial appearance refers to the distinctive face including flat nasal bridge, short upturned nose, periorbital puffiness, long philtrum, and delicate chin. SVAS refers to supravalvular aortic stenosis. Somatic cell type refers to the somatic cell type reprogrammed into iPSCs. (B) Gesell Developmental Scale data of neurocognitive, motor, language, and social assessments of the adopted three WS patients. (C) Distribution map of WS genes of Healthy CTRL and WS patients, respectively. (D) Targeted sequencing data analysis of 7q11.23 region of Healthy CTRL and WS patients, respectively.
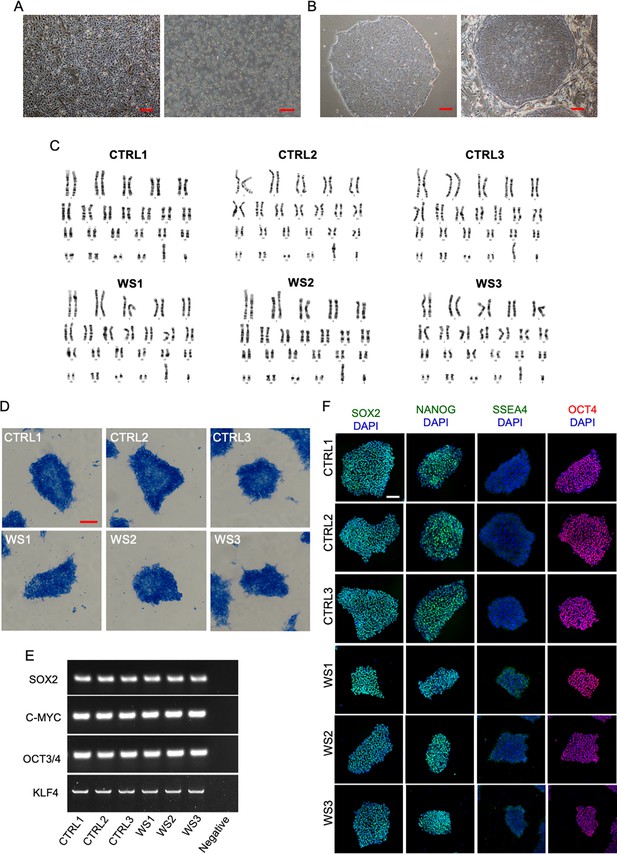
Characterization of primary cells and the generated iPSCs.
(A) Representative brightfield images of cultured urinary cells and PBMCs. Scale bars, 50 μm. (B) Representative brightfield images of cultured iPSCs. iPSCs were maintained under feeder-free condition (left). For organoid generation, iPSCs were cultured on mouse embryonic fibroblasts (feeder-dependent) (right). Scale bar, 50 μm. (C) Karyotype analysis of Healthy and WS iPSCs lines, respectively. (D) Representative images of Alkaline phosphatase staining of Healthy and WS iPSCs. Scale bar, 200 μm. (E) qRT-PCR assay results showed the expression of pluripotency factors in different iPSCs lines. Primary urinary cells were used for the negative control. (F) Representative images of immunofluorescence staining of pluripotent markers SOX2, NANOG, SSEA4, OCT4 with Healthy and WS iPSCs, respectively. Scale bar, 50 μm.
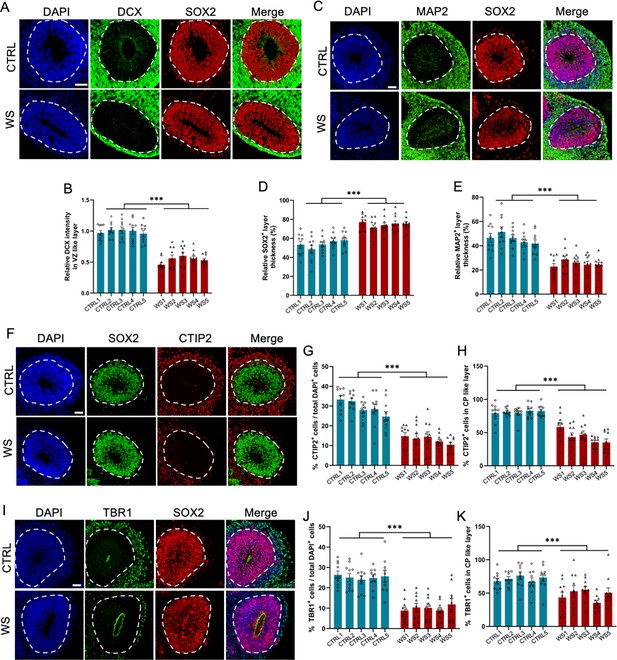
WS forebrain organoids exhibit the reduced neuronal differentiation.
(A) Representative images of DCX and SOX2 immunofluorescence staining of CTRL and WS forebrain organoids, respectively. (B) Quantification results showed that the intensity of DCX signal significantly decreased in SOX2+ VZ like layers of WS forebrain organoids compared to CTRL. (C) Representative images of MAP2 and SOX2 immunofluorescence staining of CTRL and WS forebrain organoids, respectively. (D, E) Quantification results showed that the thickness of VZ like layer (SOX2+MAP2−) significantly increased (D), but the thickness of cortical plate (CP) like layer (MAP2+) CP like layer significantly decreased (E) in WS forebrain organoids compared to CTRL. (F) Representative images of SOX2 and neuronal marker CTIP2 immunofluorescence staining of CTRL and WS forebrain organoids, respectively. (G) Quantification results showed that the proportion of CTIP2+ neurons significantly decreased in WS forebrain organoids compared to CTRL. (H) Quantification results showed that the relative thickness of SOX2+ VZ like layer significantly increased, while the the relative thickness of CTIP2+ CP like layer significantly decreased in WS forebrain organoids compared to CTRL. (I) Representative images of SOX2 and neuronal marker TBR1 immunofluorescence staining of CTRL and WS forebrain organoids, respectively. (J) Quantification results showed that the proportion of total TBR1+ neurons significantly decreased in WS forebrain organoids compared to CTRL. (K) Quantification results showed that the proportion of TBR1+ neurons significantly decreased in TBR1+ CP like layer of WS forebrain organoids compared to CTRL. For (A–K), five IPS lines of Ctrl and WS were adopted for the study, respectively. Three to five organoids were adopted from each IPS line and 10 representative rosettes of organoids were analyzed in each line. values represent mean ± SEM. *p<0.05, **p<0.01, ***p<0.001, one-way ANOVA followed by Dunnett’s multiple-comparison test. Scale bar, 50 μm.
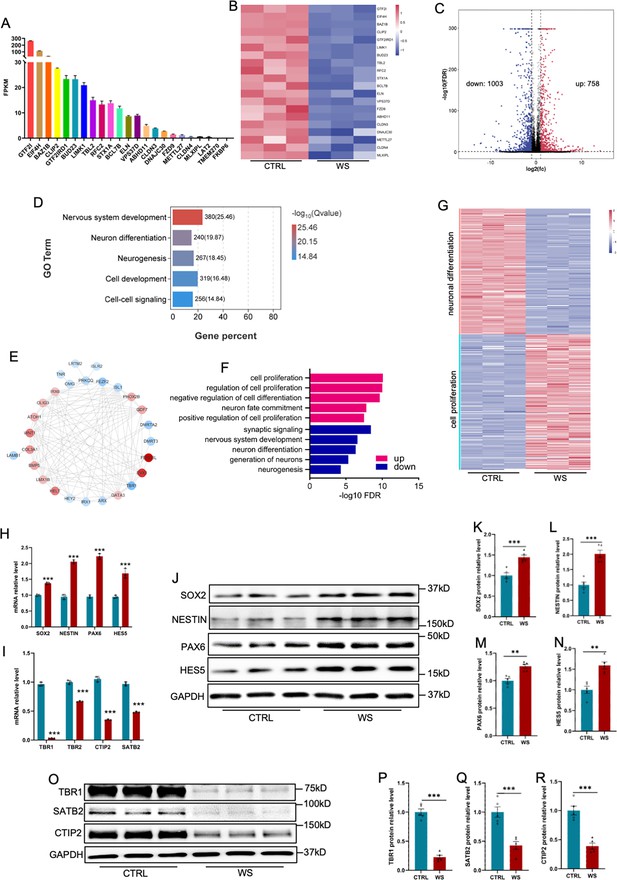
WS forebrain organoids display the altered expression of neurogenic genes.
(A) Fragments per kilobase of transcript per million mapped reads (FPKM) values of 23 WS genes in Day 56 CTRL organoids. (B) Heatmap analysis of 23 WS genes in both CTRL and WS brain organoids. (C) Volcano plot of the differentially expressed genes (DEGs). Bulk RNA sequencing (bulk RNA-seq) revealed 758 up-regulated and 1003 down-regualted genes in WS brain organoids compared with CTRL organoids, respectively. Fold change >2 and p-value <0.05 were considered as significance. (D) Gene Ontology (GO) analysis of DEGs showed the enrichment in terms related to neuronal differentiation and neurogenesis. (E) Interactome plot of top 27 genes of neurogenesis GO term was presented. Red dots representing up regulated genes and blue dots representing down regulated genes. (F) GO analysis of DEGs in WS organoids. Red bar represents terms enriched by up regulated genes, and blue bar represents terms enriched by down regulated genes. (G) Heatmap of cell proliferation and neuronal differentiation related genes in CTRL and WS organoids. (H, I) qRT-PCR results showed the increased expression of NPCs markers including SOX2, NESTIN, PAX6, and HES5 (H) and the decreased expression of neuronal markers including TBR1, TBR1, CTIP2, and SATB2 (I) in WS organoids compared to Ctrl group. Values represent mean ± SEM; *p<0.05, **p<0.01, ***p<0.001; unpaired Student’s t test. (J–N) WB assay (J) and quantification results shoed the increased expression of SOX2 (K), NESTIN (L), PAX6 (M), HES5 (N) in WS organoids compared to CTRL group. Values represent mean ± SEM; *p<0.05, **p<0.01, ***p<0.001; unpaired Student’s t test. (O–R) WB assay (O) and quantification results of TBR1 (P), CTIP2 (Q), SATB2 (R) in WS organoids compared to CTRL group. Values represent mean ± SEM; n=5 independent experiments; *p<0.05, **p<0.01, ***p<0.001; unpaired Student’s t test.
-
Figure 3—source data 1
PDF file for original blots images in Figure 3J and O.
- https://cdn.elifesciences.org/articles/98081/elife-98081-fig3-data1-v1.zip
-
Figure 3—source data 2
TIF files for original blots images in Figure 3J and O.
- https://cdn.elifesciences.org/articles/98081/elife-98081-fig3-data2-v1.zip
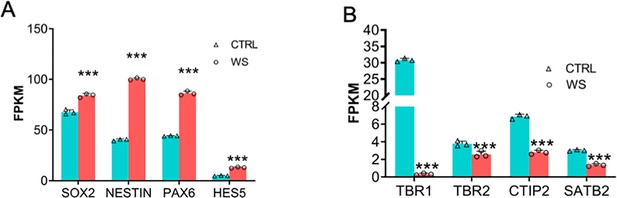
Bulk RNA sequencing (RNA-seq) reveals the altered expression of neurodevelopmental genes.
(A) FPKM values of SOX2, NESTIN, PAX6, HES5. Values represent mean ±SE; n=3 replicates for each group; *p<0.05, **p<0.01, and ***p<0.001; unpaired Student’s t test. (B) FPKM values of TBR1, TBR2, CTIP2, SATB2. Values represent mean ± SEM; n=3 replicates for each group; *p<0.05, **p<0.01, and ***p<0.001; unpaired Student’s t test.
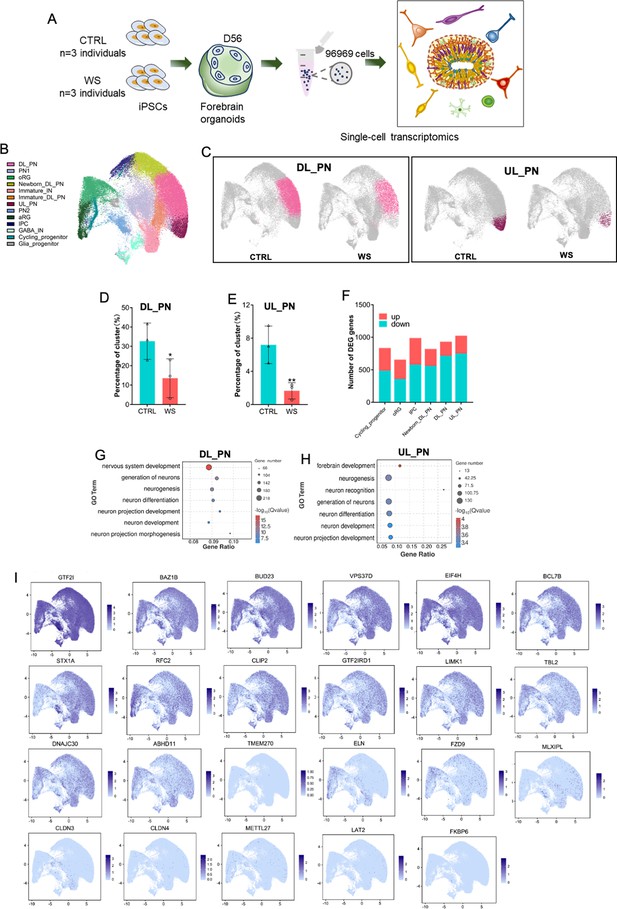
scRNA-seq reveals the abnormal neuronal differentiation in WS forebrain organoids.
(A) Schematic illustration of the scRNA-seq and histological analysis of both CTRL and WS forebrain organoids. (B) UMAP plot of cell types detected in both CTRL and WS forebrain organoids. DL_PN, deep layer projection neurons; UL_PN, upper layer projection neurons; IN, interneurons; aRG, apical radial glia; oRG, outer radial glia; IPC, intermediate progenitor cells; GABA IN, GABAergic interneurons. (C) UMAP plot of DL_PN (left) and UL_PN (right) in both CTRL and WS forebrain organoids. (D, E) Quantificationof the percentages of DL_PN and UL_PN in both CTRLcontr and WS forebrain organoids. Values represent mean ± SEM; *p<0.05, **p<0.01, ***p<0.001; unpaired Student’s t test. (F) Number of DEGs in the indicated cell types in WS organoids. Fold change >1.28 and p-value <0.05 were considered as significance. (G, H) GO analysis of DEGs in the DL_PN (I) and UL_PN (J) cell types in WS organoids. (I) UMAP visualization of the expression of WS genes in CTRL forebrain organoids.
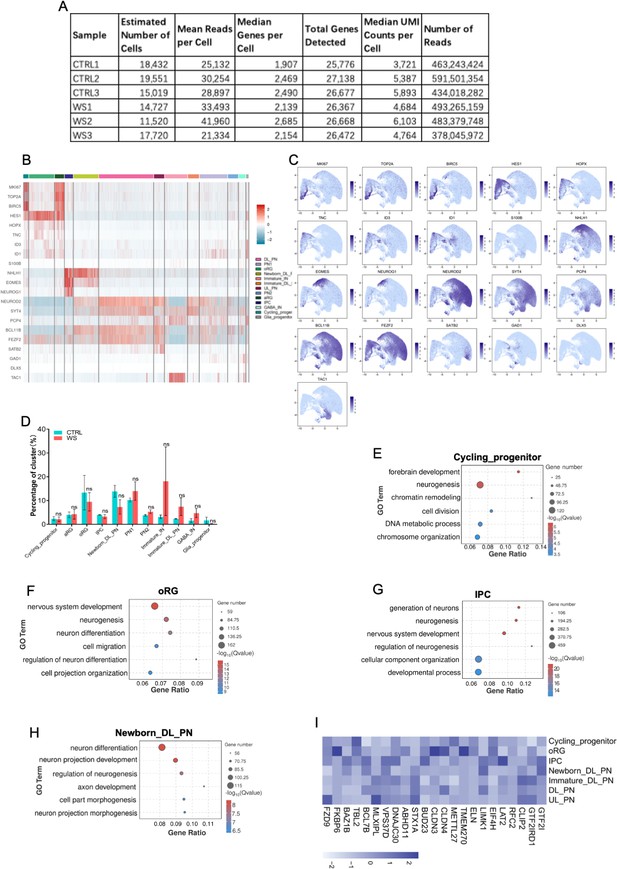
scRNA-seq reveals the abnormal neuronal differentiation in WS forebrain organoids.
(A) Statistics of the scRNA-seq of forebrain organoids from three healthy CTRL and three WS patients, respectively. (B) Heat map of marker genes for distinct cell types in both healthy CTRL and WS forebrain organoids, respectively. DL_PN, deep layer projection neurons; UL_PN, upper layer projection neurons; IN, interneuron; aRG, apical radial glia; oRG, outer radial glia; IPC, intermediate progenitor cells; GABA IN, GABAergic interneurons. (C) UMAP plots of of marker genes for distinct cell types in both healthy CTRL and WS forebrain organoids, respectively. (D) Quantification of the percentages of the indicated cell types in both healthy CTRL and WS forebrain organoids. Values represent mean ± SEM; n=3 replicates for each group; *p<0.05, **p<0.01, ***p<0.001; unpaired Student’s t test. (E–H) GO analysis of DEGs in the Cycling_progenitor (E), oRG (F), IPC (G) and newborn_DL_PN (H) cell types in WS organoids. (I) Heat map illustrating the expression of WS genes among distinct cell types in the day-56 forebrain organoids.
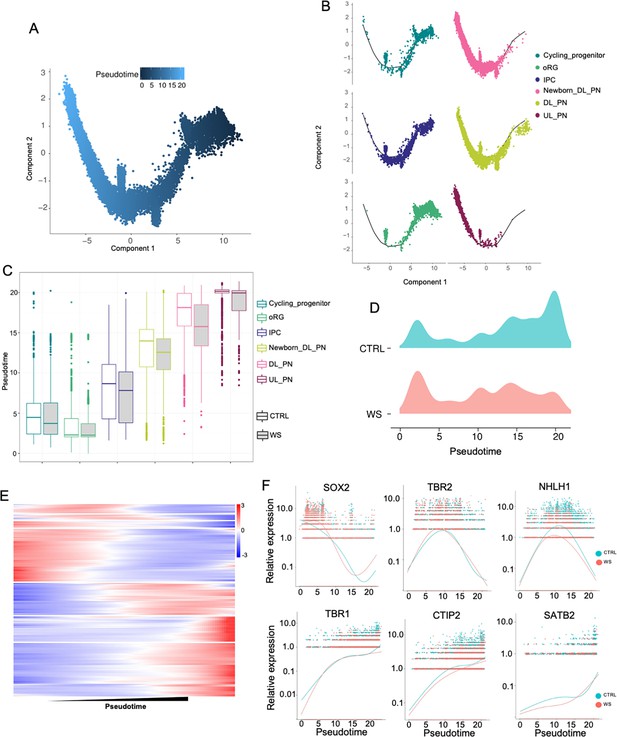
WS forebrain organoids exhibit aberrant developmental trajectory of excitatory neurons.
(A) Pseudo-time distribution of cell trajectories of excitatory neurons related cell clusters. (B) Distribution of cell cluster in Pseudotime trajectories. (C) Boxplot showing the distribution of pseudotime within each cell type in both CTRL and WS-derived forebrain organoids. (D) Mountain maps of pseudotime uniform manifold approximation in both CTRL and WS forebrain organoids. (E) Heatmap of pseudotime line-dependent gene expression. (F) Expression of SOX2,TBR2, NHLH1, TBR1, CTIP2 and SATB2 ordered by Monocle analysis in pseudotime.
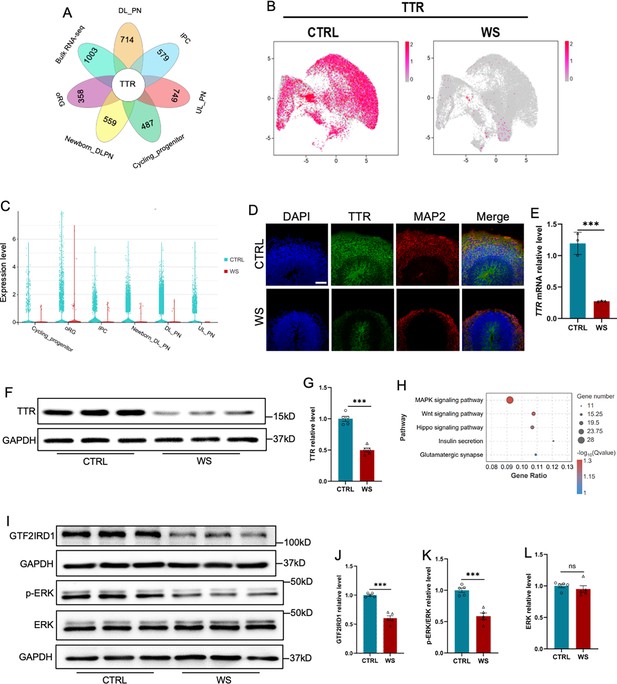
The deficiency of GTF2IRD1 leads to the down-regualtion of Transthyretin and inhibition of ERK signaling.
(A) Integrated analysis of down-regualted genes in 6 clusters relating to the generation of excitatory neurons and down-regualted genes revealed by bulk RNA-seq. (B) UMAP visualization of the expression of TTR in CTRL and WS forebrain organoids, respectively. (C) Transthyretin (TTR) displayed the decreased expression in 6 clusters relating to the generation of excitatory neurons. (D) Representative images of TTR immunofluorescence staining with in CTRL and WS forebrain organoids, respectively. Scale bar, 100 μm. (E) qRT-PCR results showed that TTR level significantly decreased in WS forebrain organoids compared to Ctrl group. Values represent mean ± SEM; n=3 independent experiments; *p<0.05, **p<0.01, ***p<0.001; unpaired Student’s t test. (F, G) WB assay (F) and quantification results (G) showed that the level of TTR was significantly decreased in WS brain organoids compared to Ctrl. Values represent mean ± SEM; n=5 independent experiments; *p<0.05, **p<0.01, ***p<0.001; unpaired Student’s t test. (H) KEGG analysis showed that down-regualted genes enriched for MAPK pathway, etc. (I–K) WB assay (I) and quantification results showed that the level of phosphorylated ERK (p-ERK) was significantly decreased (J), but the level of total ERK (K) was not changed in WS brain organoids compared to Ctrl. Values represent mean ± SEM; n=5 independent experiments; *p<0.05, **p<0.01, ***p<0.001; unpaired Student’s t test. (L, M) WB assay (L) and quantification results (M) showed that the level of GTF2IRD1 was significantly decreased in WS brain organoids compared to Ctrl. Values represent mean ± SEM; n=5 independent experiments; *p<0.05, **p<0.01, ***<0.001; unpaired Student’s t test.
-
Figure 6—source data 1
PDF file for original blots images in Figure 6F and I.
- https://cdn.elifesciences.org/articles/98081/elife-98081-fig6-data1-v1.zip
-
Figure 6—source data 2
TIF files for original blots images in Figure 6F and I.
- https://cdn.elifesciences.org/articles/98081/elife-98081-fig6-data2-v1.zip
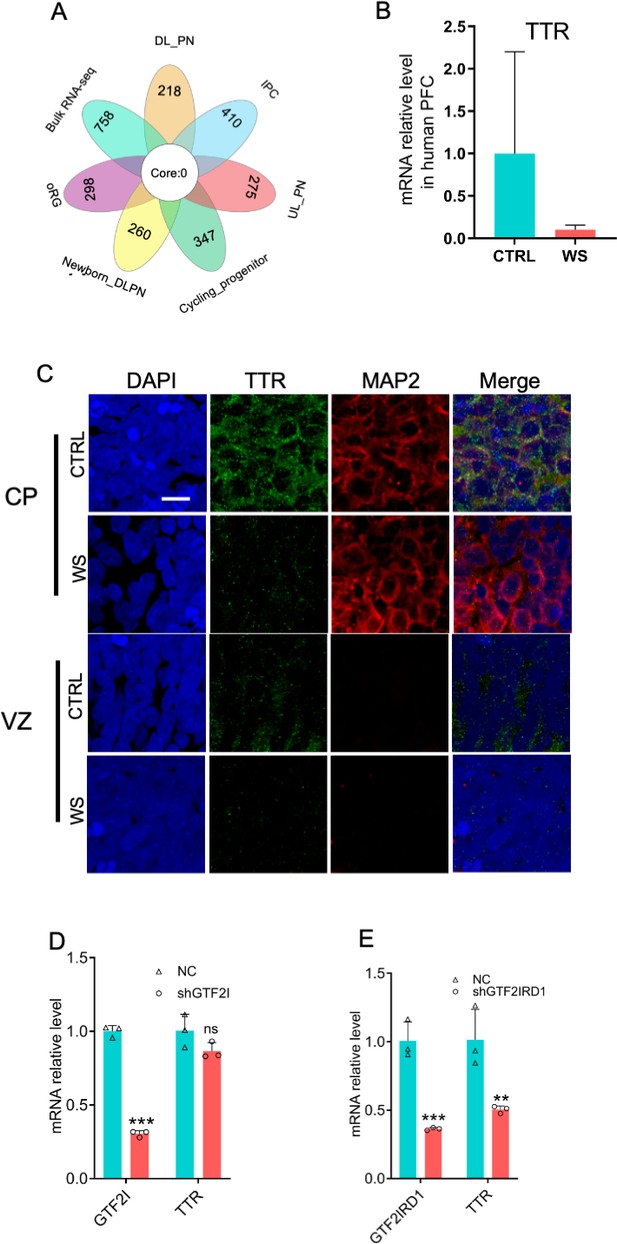
The level of TTR decreases in WS organoids.
(A) Integrated analysis of up-regualted genes in 6 clusters relating to the generation of excitatory neurons and up-regualted genes revealed by bulk RNA-seq. (B) mRNA level of TTR was decreased in the prefrontal cortex of WS patient compared to CTRL. This analysis based on the published data generated by Barak et al., 2019. (C) Representative images of TTR staining showed that TTR was mainly expressed in CP region of CTRL organoids, and the signal intensity remarkably decreased in WS organoids. Scale bar, 100 μm. (D) qRT-PCR results showed that GTF2I knockdown showed no effect on the expression of TTR in HEK293T cells. Values represent mean ± SEM; n=3 independent experiments; *p<0.05, **p<0.01, **p<0.001; unpaired Student’s t test. (E) qRT-PCR results showed that GTF2IRD1 knockdown significantly reduced the expression of TTR in HEK293T cells. Values represent mean ± SEM; n=3 independent experiments; *p<0.05, **p<0.01, ***p<0.001; unpaired Student’s t test.
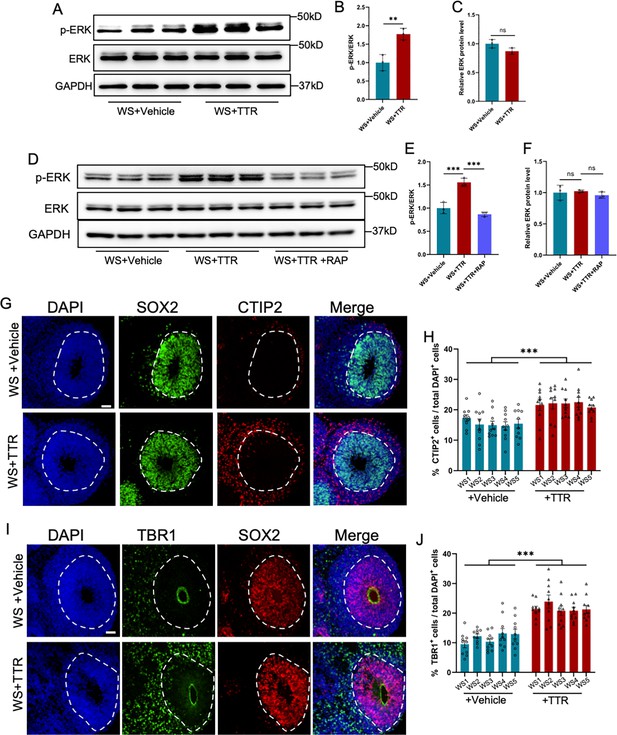
Recombinant TTR rescues neurodevelopmental defects of WS organoids.
(A–C) WB assay (A) and quantification results showed that TTR exposure enhanced the level of p-ERK in WS brain organoids (B), while the level of total ERK was not affected (C). Values represent mean ± SEM; n=3; *p<0.05, **p<0.01, ***p<0.001; unpaired Student’s t test. (D–F) Western blot (D) and quantification results showed that TTR exposure enhanced the level of p-ERK (E), which could be inhibited by receptor-associated protein (RAP) in WS brain organoids (F). Values represent mean ± SEM; n=3 independent experiments; *p<0.05, **p<0.01, ***p<0.001; unpaired Student’s t test. (G) Representative images of SOX2 and CTIP2 immunofluorescence staining with WS-derived forebrain organoids under different culture conditions. Scale bar, 50 μm. (H) Quantification results showed that TTR exposure significantly increased the proportion of CTIP2+ neurons in WS-derived forebrain organoids compared to CTRL. Values represent mean ± SEM; n=30–35 VZ-like regions of 3–5 organoids from three WS patients in each group. *p<0.05, **p<0.01, ***p<0.001; unpaired Student’s t test. (I) Representative images of SOX2 and TBR1 immunofluorescence staining with WS-derived forebrain organoids under different culture conditions. Scale bar, 50 μm. (J) Quantification results showed that TTR exposure significantly increased the proportion of TBR1+ neurons in WS-derived forebrain organoids. Values represent mean ± SEM; n=30–35 VZ-like regions of 3–5 organoids from three WS patients in each group. *p<0.05, **p<0.01, ***p<0.001; unpaired Student’s t test.
-
Figure 7—source data 1
PDF file for original blots images in Figure 7A and D.
- https://cdn.elifesciences.org/articles/98081/elife-98081-fig7-data1-v1.zip
-
Figure 7—source data 2
TIF files for original blots images in Figure 7A and D.
- https://cdn.elifesciences.org/articles/98081/elife-98081-fig7-data2-v1.zip
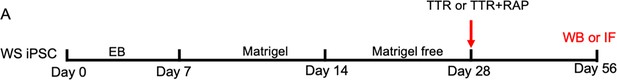
Schematic illustration of exogenous TTR application.
(A) Schematic of the application procedure of recombinant TTR, and TTR plus receptor-associated protein (RAP, one TTR inhibitor) for WS forebrain organoid. TTR or TTR +RAP was supplemented to brain organoids at day 28, and assays were performed at day 56.
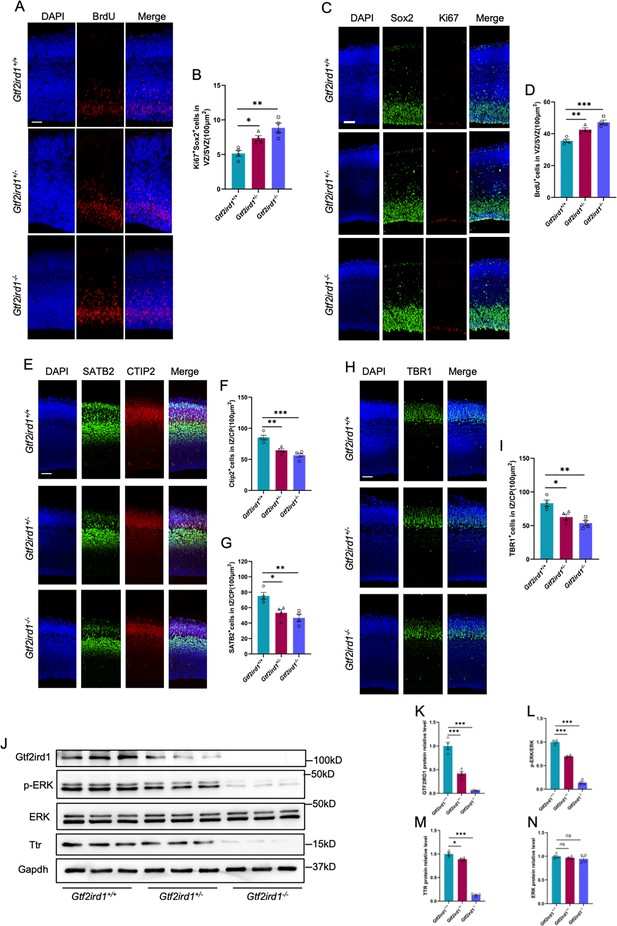
Gtf2ird1 deficient mice display neurodevelopmental deficits and the reduced TTR.
(A, B) Representative images of BrdU immunofluorescence staining (A) and quantification results (B) showed the increased BrdU+ eNPCs in VZ/SVZ of Gtf2ird1+/- (heterozygous, Het) and Gtf2ird1-/- (knockout, KO) mice compared to Gtf2ird1+/+ (Wild-type, WT) mice, respectively. Pups were injected with BrdU at E15.5 and sacrificed 1.5 hr later. Values represent mean ± SEM; n=4 animals for each group; ∗p<0.05, ∗∗p<0.01, ∗∗∗p<0.001; unpaired Student’s t test. Scale bar, 50 μm. (C, D) Representative images of Sox2 and Ki67 immunofluorescence staining (C) and quantification results (D) showed the increased Ki67+Sox2+ eNPCs in VZ/SVZ of Het and KO mice compared to WT mice, respectively. Values represent mean ± SEM; n=4 animals for each group; *p<0.05, **p<0.01, ***p<0.001; unpaired Student’s t test. Scale bar, 50 μm. (E–G) Representative images of neuronal marker SATB2 and CTIP2 immunofluorescence staining (E) and quantification results showed the decreased Ctip2+ (F) and SATB2+ (G) neurons of Het and KO mice compared to WT mice, respectively. Values represent mean ± SEM; n=4 animals for each group; *p<0.05, **p<0.01, ***p<0.001; unpaired Student’s t test. Scale bar, 50 μm. (H, I) Representative images of neuronal marker Tbr1 immunofluorescence staining (H) and quantification results (I) showed the decreased Tbr1+ neurons in IZ/CP of Het and KO mice compared to WT mice, respectively. Values represent mean ± SEM; n=4 animals for each group; *p<0.05, **p<0.01, ***p<0.001; unpaired Student’s t test. Scale bar, 50 μm. (J–N) WB assay (J) and quantification results showed that the levels of Gtf2ird1 (K), p-ERK (L) and TTR (M) were significantly decreased, while the level of total ERK (N) was not altered in the cortical tissues of Het and KO mice compared to WT mice, respectively. Values represent mean ± SEM; n=4 animals for each group; *p<0.05, **p<0.01, ***p<0.001; unpaired Student’s t test.
-
Figure 8—source data 1
PDF file for original blots images in Figure 8J.
- https://cdn.elifesciences.org/articles/98081/elife-98081-fig8-data1-v1.zip
-
Figure 8—source data 2
TIF files for original blots images in Figure 8J.
- https://cdn.elifesciences.org/articles/98081/elife-98081-fig8-data2-v1.zip
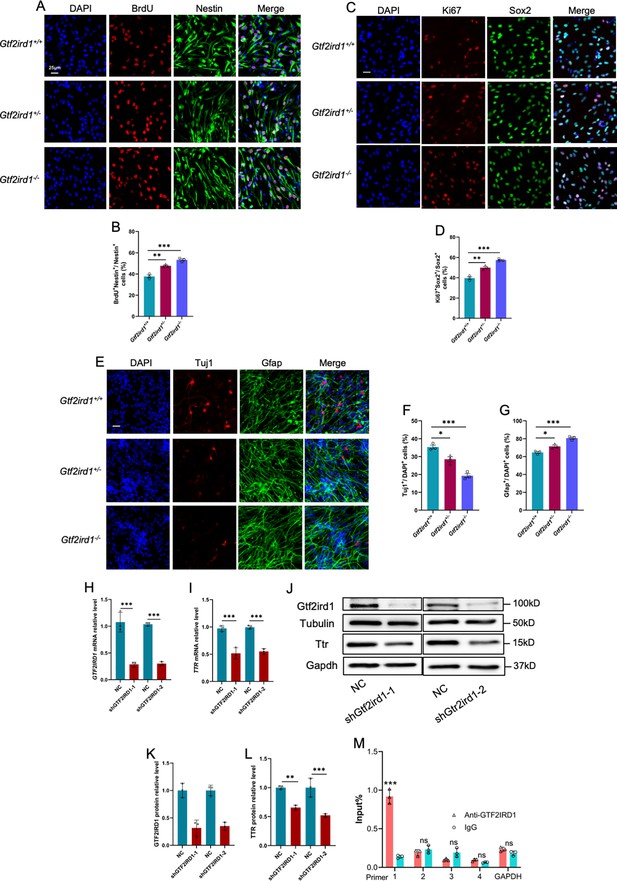
Gtf2ird1 regulates the proliferation and differentiation of eNPCs in vitro.
(A) Representative images of BrdU and Nestin immunofluorescence staining with cultured eNPCs from WT, Het and KO mice, respectively. Scale bar, 25 μm. (B) Quantification results showed that the percentage of BrdU+/BrdU+Nestin+ eNPCs was significantly increased in Het and KO mice relative to WT cells, respectively. Values represent mean ± SEM; n=3 independent experiments; *p<0.05, **p<0.01, ***p<0.001; unpaired Student’s t test. (C) Representative images of Ki67 and Sox2 immunofluorescence staining with cultured eNPCs from WT, Het and KO mice, respectively. Scale bar, 25 μm. (D) Quantification results showed that the percentage of Ki67+/Ki67+Sox2+ eNPCs was significantly increased in Het and KO mice relative to WT cells, respectively. Values represent mean ± SEM; n=3 independent experiments; *p<0.05, **p<0.01, ***p<0.001; unpaired Student’s t test. (E) Representative images of Tuj1 and Gfap immunofluorescence staining with cultured eNPCs from WT, Het, and KO mice, respectively. Scale bar, 25 μm. (F, G) Quantification results showed that the nubmer of Tuj1+ neurons was significantly decreased (F), but the number of Gfap +astrocytes was significantly increased (G) upon the differentiation of eNPCs in Het and KO mice relative to WT cells, respectively. Values represent mean ± SEM; n=3 independent experiments; *p<0.05, **p<0.01, ***p<0.001; unpaired Student’s t test. (H, I) qRT-PCR results showed that acute knockdown of Gtf2ird1 significantly reduced the level of Gtf2ird1 (H) and TTR (I) in WT eNPCs compared to scramble group (NC), respectively. Values represent mean ± SEM; n=3 independent experiments; *p<0.05, **p<0.01, ***p<0.001; unpaired Student’s t test. (J–L) WB assay (J) and quantification results showed that acute knockdown of Gtf2ird1 significantly reduced the level of Gtf2ird1 (K) and TTR (L) in WT eNPCs, respectively. Values represent mean ± SEM; n=3 independent experiments; *p<0.05, **p<0.01, ***p<0.001; unpaired Student’s t test. (M) Chromatin immunoprecipitation followed qPCR (ChIP-qPCR) showed that Gtf2ird1 binds to the promoter regions of TTR. Values represent mean ± SEM; n=3 independent experiments; *p<0.05, **p<0.01, ***p<0.001; unpaired Student’s t test.
-
Figure 9—source data 1
PDF file for original blots images in Figure 9J.
- https://cdn.elifesciences.org/articles/98081/elife-98081-fig9-data1-v1.zip
-
Figure 9—source data 2
TIF files for original blots images in Figure 9J.
- https://cdn.elifesciences.org/articles/98081/elife-98081-fig9-data2-v1.zip
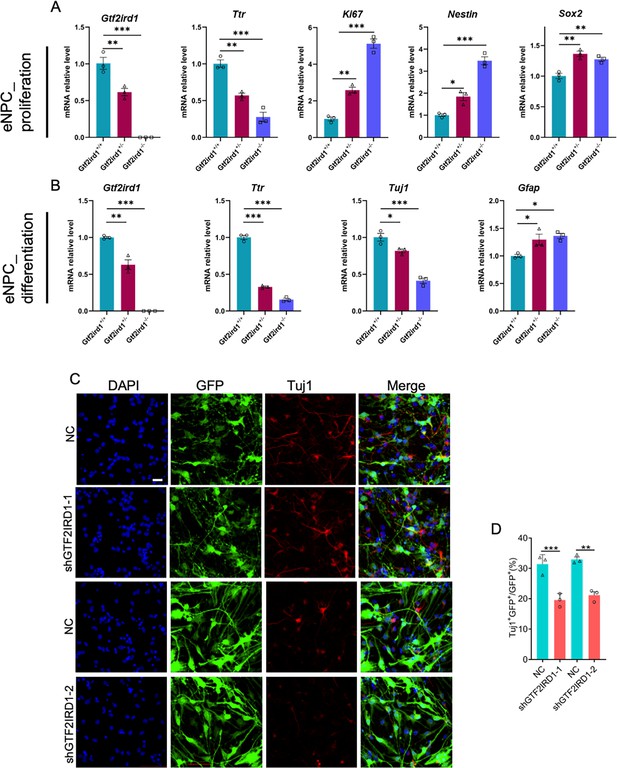
Gtf2ird1 deficiency induces the abnormal proliferation and differentiation of embryonic neuronal progenitor cells in vitro.
(A) qRT-PCR results showed the decreased levels of Gtf2ird1 and Ttr, but the increased levels of Ki67, Nestin and Sox2 in proliferating Het and KO embryonic neuronal progenitor cells (eNPCs) relative to WT cells. Values represent mean ± SEM; n=3 independent experiments; *p<0.05, **p<0.01, ***p<0.001; unpaired Student’s t test. (B) qRT-PCR results showed the decreased Gtf2ird1, Ttr and Tuj1, but the increased Gfap in differentiated Het and KO eNPCs relative to WT cells. Values represent mean ± SEM; n=3 independent experiments; *p<0.05, **p<0.01, ***p<0.001; unpaired Student’s t test. (C, D) Representative images (C) and quantification results (D) showed that Gtf2ird1 knockdown led to a significant decrease of new born neurons in mouse eNPCs. Values represent mean ± SEM; n=3 independent experiments; *p<0.05, **p<0.01, ***p<0.001; unpaired Student’s t test. Scale bar, 20 μm.
Additional files
-
Supplementary file 1
DEGs in WS brain organoids.
- https://cdn.elifesciences.org/articles/98081/elife-98081-supp1-v1.xlsx
-
Supplementary file 2
Information of reagents.
- https://cdn.elifesciences.org/articles/98081/elife-98081-supp2-v1.xlsx
-
Supplementary file 3
List of primers.
- https://cdn.elifesciences.org/articles/98081/elife-98081-supp3-v1.pdf
-
MDAR checklist
- https://cdn.elifesciences.org/articles/98081/elife-98081-mdarchecklist1-v1.docx