FGF8-mediated gene regulation affects regional identity in human cerebral organoids
Figures
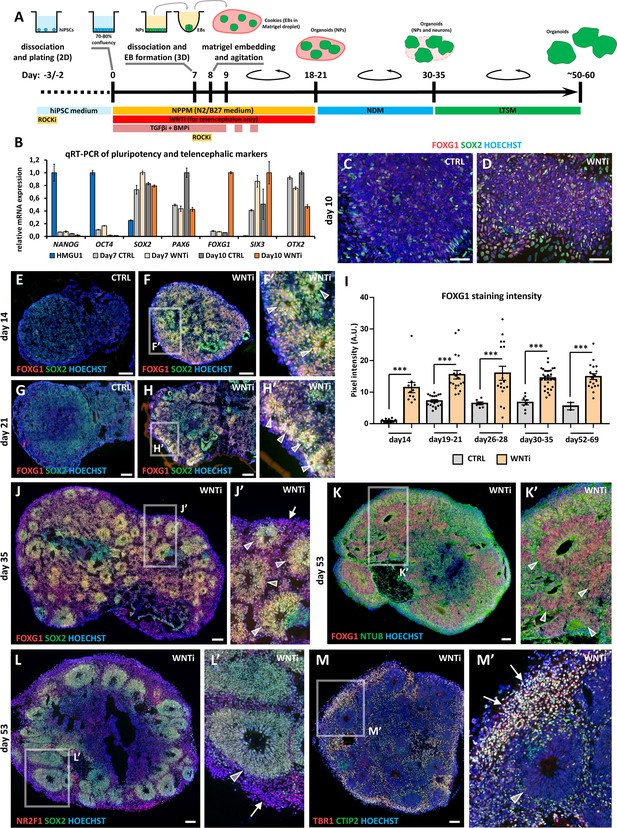
Hybrid 2D/3D protocol for fast and reproducible generation of human cortical organoids.
(A) Schematic of the hybrid 2D/3D method for generating telencephalic/cortical human organoids in vitro, using a triple inhibition of TGFβ, BMP, and WNT pathways (SB-431542, 5 µM; LDN-193189, 0.25 µM; XAV-939, 2 µM). On day 7, cells are dissociated and re-aggregated in 96-well plates. One day later, embryoid bodies (EBs) are embedded in Matrigel droplets (10 µL per droplet containing 1–4 EBs). These droplets, termed ‘cookies’, are then cultured in spinning bioreactors. (B) Real-time qRT-PCR analysis quantifying pluripotency markers (NANOG, OCT4) and telencephalic neural progenitor (NP) markers (SOX2, PAX6, FOXG1, SIX3, and OTX2) in undifferentiated HMGU1 hiPSCs and in day7 and day10 control (CTRL) and WNT-inhibited (WNTi) samples, as indicated. n=2 culture wells per condition, pooled prior to RNA extraction. (C,D) Immunostaining for FOXG1 (red) and SOX2 (green) in day10 2D neural cultures under control (CTRL) conditions (C) or following WNT inhibition (WNTi) (D). (E–I) Immunostaining for FOXG1 (red) and SOX2 (green) in day14 (E-F’) and day21 (G-H’) organoids under CTRL or WNTi conditions, as indicated. White arrowheads in high-magnification images point to neural progenitor (NP) rosettes. The graph (I) shows quantification of FOXG1 pixel intensity in CTRL and WNTi samples across time points. n≥7 sections from n≥4 organoids from n=2 independent batches (except day52-69 CTRL sample, n=2 sections from 1 batch). (J,J’) FOXG1 (red) and SOX2 (green) immunostaining in day35 WNTi organoids. White arrowheads in high-magnification images indicate NP neural rosettes, while arrows highlight differentiating neurons surrounding the rosettes. (K-M’) Immunostaining for FOXG1 (red) and NTUB (green) (K, K’), NR2F1 (red) and SOX2 (green) (L, L’), and TBR1 (red) and CTIP2 (green) (M, M’) in day53 WNTi organoids. High-magnification images highlight FOXG1+ SOX2+ NR2F1+ NP rosettes/neuroepithelia (K-L’; indicated by white arrowheads) surrounded by TBR1+ CTIP2+ NR2F1+ differentiating cortical neurons (L’-M’; indicated by white arrows). Scale bars: 100 µm.
-
Figure 1—source data 1
Expression level of pluripotency and telencephalic markers in 2D human progenitors.
Raw quantitative RT-PCR data showing telencephalic and pluripotency marker expression in control and WNTi cells on days 7 and 10.
- https://cdn.elifesciences.org/articles/98096/elife-98096-fig1-data1-v2.xls
-
Figure 1—source data 2
FOXG1 staining intensity in CTRL and WNTi human organoids.
GraphPad sheet containing raw pixel intensity data measured in CTRL and WNTi organoid cryostat slices from day14 to day69 following FOXG1 immunostaining.
- https://cdn.elifesciences.org/articles/98096/elife-98096-fig1-data2-v2.zip

Hybrid 2D/3D protocol for generation of telencephalic human organoids.
Brightfield microscope images showing, from top left to bottom right: undifferentiated and confluent HMGU1 cells at the starting day of the neural differentiation protocol; day7 neural progenitors organized in groups of radially oriented cells (arrowheads) and surrounded by non-neural cells; dissociated NPs at day7, spinned at the bottom of 96-well plates; day7 dissociated NPs at the bottom of a single well; embryoid body (EB) at day8, 24 hr after dissociation; day10 EB included in Matrigel; EB at day16, with some rosettes and neural epithelia becoming visible on the borders; day65 organoid.
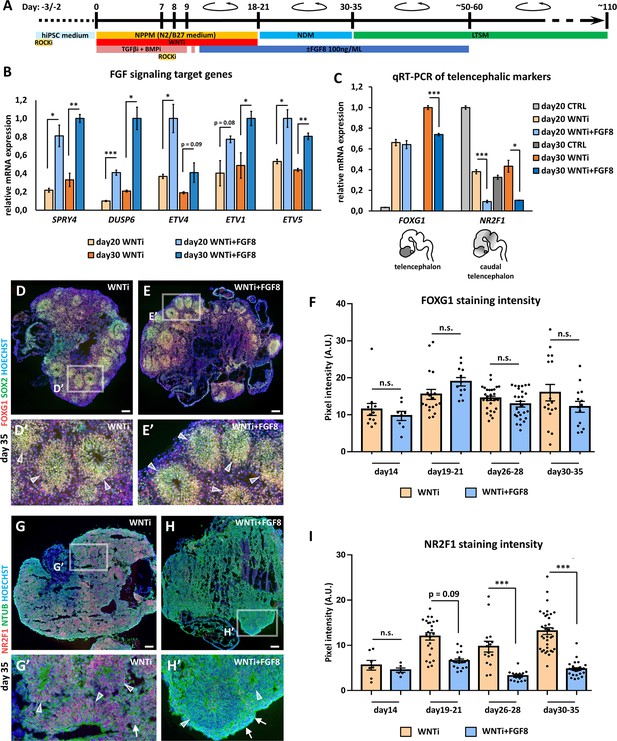
FGF8-mediated regulation of target gene expression in FOXG1+ telencephalic organoids.
(A) Schematic of the hybrid 2D/3D method for applying FGF8 treatment on telencephalic/cortical human organoids in vitro. FGF8 (100 ng/mL) was added to the neural progenitor patterning medium (NPPM) beginning on day10-11 (blue bar) and maintained through subsequent culture steps until approximately day50-60. (B) Real-time qRT-PCR analysis of FGF8 target gene expression (SPRY4, DUSP6, ETV4, ETV1, and ETV5) in day20 and day30 organoids treated with WNT inhibition alone (WNTi) or in combination with FGF8 (WNTi + FGF8), as indicated. n=3 organoids per condition, pooled prior RNA extraction. (C) Real-time qRT-PCR quantification of FOXG1 (telencephalic marker) and NR2F1 (caudal telencephalic marker and FGF8 target) expression in day20 and day30 control (CTRL), WNT-inhibited (WNTi), and FGF8-treated (WNTi + FGF8) samples, as indicated. FGF8 treatment effectively downregulates NR2F1 expression in WNTi + FGF8 organoids compared with WNTi organoids. n=3 organoids per condition, pooled prior RNA extraction. (D–F) Immunostaining for FOXG1 (red) and SOX2 (green) in day35 WNTi and WNTi + FGF8 organoids, as indicated. FGF8 treatment does not significantly alter FOXG1 expression. White arrowheads in high-magnification images indicate SOX2+ NR2 F1+ NPs within rosettes. Graph (F) shows pixel intensity quantification of FOXG1 staining in WNTi and WNTi + FGF8 organoids at different time points. n≥8 sections from n≥4 organoids from n≥2 distinct batches. (G–I) NR2F1 and NTUB (red and green, respectively, in G-H’) immunostainings on day35 WNTi and WNTi + FGF8 organoids, as indicated. FGF8 treatment efficiently modulates NR2F1 expression (compare G and H). High-magnification images (G’ and H’) show neural rosettes (NTUBlow, indicated by white arrowheads) and differentiating neurons (NTUBhigh, indicated by white arrows), both expressing NR2F1 (red) in WNTi organoids, but lacking NR2F1 in WNTi + FGF8 organoids. Graph (I) displays pixel intensity quantification of NR2F1 staining in WNTi and WNTi + FGF8 organoids over time. n≥6 sections from n≥4 organoids from n≥2 distinct batches. Scale bars: 100 µm.
-
Figure 2—source data 1
Quantitative RT-PCR data for FGF target genes in human organoids.
Raw data of FGF target gene expression in day 20 and day 30 CTRL, WNTi, and WNTi +FGF8 organoids.
- https://cdn.elifesciences.org/articles/98096/elife-98096-fig2-data1-v2.xls
-
Figure 2—source data 2
Quantitative RT-PCR data for FOXG1 and NR2F1 in human organoids.
Raw data showing FOXG1 and NR2F1 expression levels in day 20 and day 30 CTRL, WNTi, and WNTi + FGF8 organoids.
- https://cdn.elifesciences.org/articles/98096/elife-98096-fig2-data2-v2.xls
-
Figure 2—source data 3
FOXG1 staining intensity in WNTi and WNTi + FGF8 human organoids.
GraphPad sheet with raw pixel intensity data from day 14 to day 35, measured in WNTi and WNTi + FGF8 organoid cryostat slices following FOXG1 immunostaining.
- https://cdn.elifesciences.org/articles/98096/elife-98096-fig2-data3-v2.zip
-
Figure 2—source data 4
NR2F1 staining intensity in WNTi and WNTi + FGF8 human organoids.
GraphPad sheet with raw pixel intensity data from day 14 to day 35, measured in WNTi and WNTi + FGF8 organoid cryostat slices following NR2F1 immunostaining.
- https://cdn.elifesciences.org/articles/98096/elife-98096-fig2-data4-v2.zip
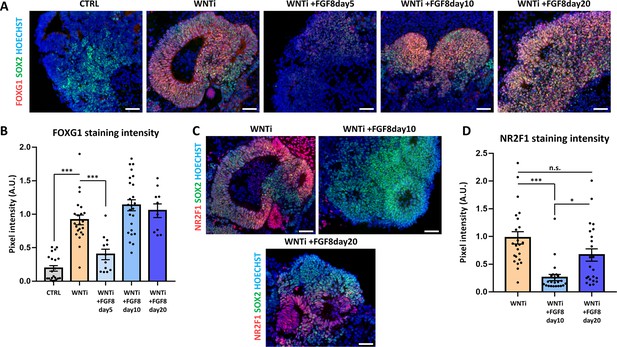
Effect of early or late FGF8 treatment on FOXG1 and NR2F1 expression in human organoids.
(A,B) Panel showing FOXG1 (red) and SOX2 (green) immunostaining in control (CTRL), WNT inhibited (WNTi) and FGF8 treated (WNTi + FGF8) day 30 organoids, as indicated. FGF8 was added at day5, at day10 or at day20; when added at day5, FGF8 partially inhibited FOXG1 expression, suggesting interference with induction of telencephalic identity (pixel intensity in graph B). (C,D) Panel showing NR2F1 (red) and SOX2 (green) immunostaining in WNT inhibited (WNTi) and FGF8 treated (WNTi + FGF8) day 30 organoids, as indicated. FGF8 treatment starting at day10 efficiently downregulated NR2F1, while treatment starting at a later time point (day20) failed to restrain NR2F1 expression (pixel intensity in graph D). For both graphs in B and D: n≥8 sections from n≥4 organoids from n=1 batch. Scale bars: 50 µm.
-
Figure 2—figure supplement 1—source data 1
FOXG1 staining intensity following early or late FGF8 treatment.
GraphPad sheet with raw pixel intensity data measured in CTRL, WNTi and WNTi + FGF8 organoids after FOXG1 immunostaining.
- https://cdn.elifesciences.org/articles/98096/elife-98096-fig2-figsupp1-data1-v2.zip
-
Figure 2—figure supplement 1—source data 2
NR2F1 staining intensity following early or late FGF8 treatment.
GraphPad sheet containing NR2F1 pixel intensity data for WNTi organoids and organoids treated with FGF8 at two distinct time points.
- https://cdn.elifesciences.org/articles/98096/elife-98096-fig2-figsupp1-data2-v2.zip
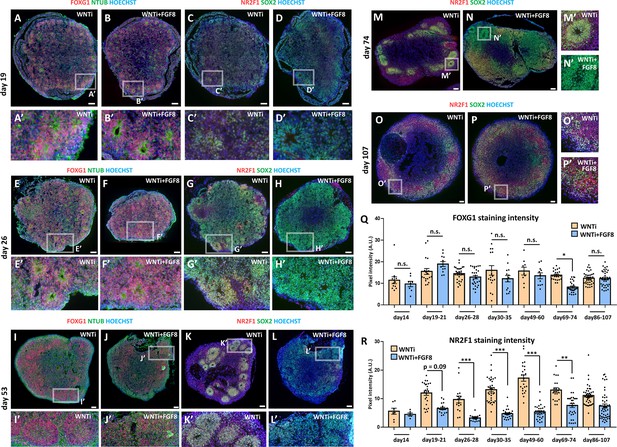
FGF8-mediated control of NR2F1 level in FOXG1+ telencephalic organoids.
(A-H’) FOXG1 and NTUB (red and green, respectively, in A-B’ and in E-F’) and NR2F1 and SOX2 (red and green, respectively, in C-D’ and in G-H’) immunostainings on day19 or on day26 WNTi and WNTi + FGF8 organoids, as indicated. FGF8 treatment did not significantly interfere with FOXG1 expression (A-B’, E-F’), while NR2F1 (which started to be detectable at low levels by immunostaining at day19 in WNTi organoids) was down-regulated by FGF8 (compare C,C’ with D,D’ or G,G’ with H,H’). (I-L’) FOXG1 and NTUB (red and green, respectively, in I-J’) and NR2F1 and SOX2 (red and green, respectively, in K-L’) immunostainings on day53 WNTi and WNTi + FGF8 organoids, as indicated. (M-P’) NR2F1 (red) and SOX2 (green) immunostaining in day74 (M-N’) and in day107 (O-P’) WNTi and WNTi + FGF8 organoids, showing that NR2F1 is still efficiently modulated by FGF8 20–25 days after end of the treatment (N, N’) but is gradually upregulated back to control levels in long-term cultured organoids (day107; P-P’). (Q) Graph shows pixel intensity quantification of FOXG1 levels after immunostaining of WNTi and WNTi + FGF8 organoids at different time points, as indicated. n≥8 sections from n≥4 organoids from n≥1 batch. (R) Graph shows pixel intensity quantification of NR2F1 immunostaining in WNTi and WNTi + FGF8 organoids at different time points, as indicated. NR2F1 level was efficiently downregulated by FGF8 treatment from day26 to day74, while it raised back at later time points. n≥6 sections from n≥4 organoids from n≥1 batch. Scale bars: 100 µm.
-
Figure 2—figure supplement 2—source data 1
FOXG1 staining intensity in WNTi and WNTi + FGF8 human organoids.
GraphPad sheet with raw pixel intensity data measured in WNTi and WNTi + FGF8 organoids following FOXG1 immunostaining at different stages.
- https://cdn.elifesciences.org/articles/98096/elife-98096-fig2-figsupp2-data1-v2.zip
-
Figure 2—figure supplement 2—source data 2
NR2F1 staining intensity in WNTi and WNTi + FGF8 human organoids.
GraphPad sheet with raw pixel intensity data measured in WNTi and WNTi +FGF8 organoids following NR2F1 immunostaining at different stages.
- https://cdn.elifesciences.org/articles/98096/elife-98096-fig2-figsupp2-data2-v2.zip
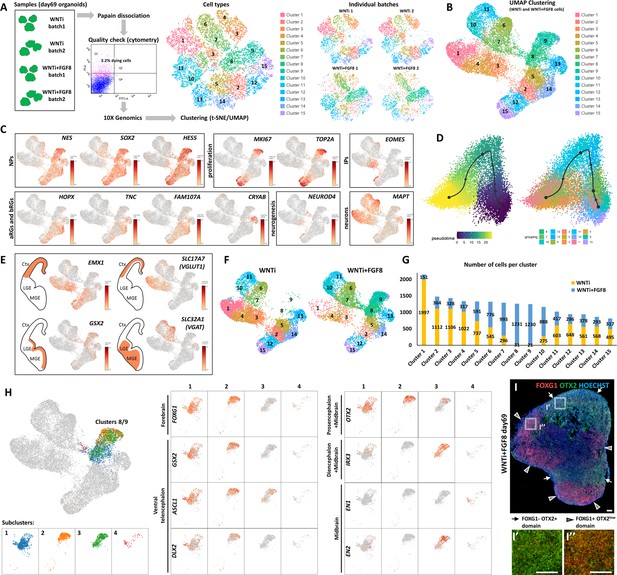
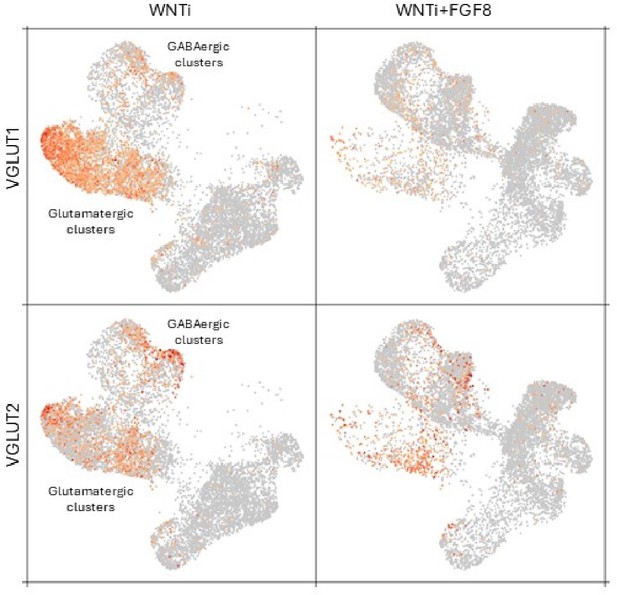
Single-cell RNA sequencing (scRNAseq) analysis of FGF8-induced cellular and molecular changes in human organoids.
(A) Experimental setup for scRNA-seq analysis of control (WNTi) and FGF8-treated (WNTi + FGF8) telencephalic organoids at day 69. Two independent batches of WNTi and WNTi + FGF8 organoids (each containing 2–3 organoids) were dissociated into single cells and processed using Chromium (10 X Genomics technology). Cells were clustered and visualized in 2D space using t-SNE and UMAP algorithms. (B) UMAP clustering of WNTi and WNTi + FGF8 cells, identifying 15 distinct clusters. (C) Expression levels of known markers for different cell types, including neural progenitor cells (NPs: NESTIN, SOX2, HES5), proliferating progenitors (KI67, TOP2A), intermediate progenitors (IPs: EOMES), apical and basal radial glia cells (aRGs and bRGs: HOPX, TNC, FAM107A, CRYAB), and differentiating/differentiated neurons (NEUROD4 and MAPT, respectively). (D) Trajectory analysis showing the most probable developmental progression from NP clusters (2, 5, 8, 9, 12, 15) to post-mitotic cell types (notably clusters 1, 3, 4, 6, 7). (E) Expression level and cluster distribution of dorsal glutamatergic markers EMX1 and SLC17A7 (also called VGLUT1) and ventral GABAergic markers GSX2 and SLC32A1 (also called VGAT), indicating the coexistence of both glutamatergic and GABAergic NPs and neurons within FOXG1+ telencephalic organoids. (F, G) UMAP clustering of WNTi and WNTi + FGF8 cells shown separately, illustrating 15 distinct clusters and their respective proportions in each condition. Panel (G) shows the number of cells in each cluster originating from WNTi (yellow) or WNTi + FGF8 (blue) organoids. (H) UMAP projection of day69 organoid scRNA-seq data, identifying four cellular groups through sub-clustering analysis on WNTi + FGF8 clusters 8 and 9. Center and right panels display expression levels of markers for the forebrain (FOXG1), ventral telencephalon (GSX2, ASCL1 and DLX2), forebrain/midbrain (OTX2), diencephalon/mesencephalon (IRX3), and mesencephalon (EN1, EN2) across the four sub-clusters. (I-I’’) Immunostaining for FOXG1 (red) and OTX2 (green) in day69 WNTi + FGF8 organoids, showing distinct FOXG1+ and FOXG1- regions. White arrows indicate FOXG1- OTX2+ non-telencephalic areas (high magnification in I’), while arrowheads denote FOXG1+ OTX2low telencephalic areas (high magnification in I’’). Ctx, cortex; MGE, medial ganglionic eminence; LGE, lateral ganglionic eminence.
-
Figure 3—source data 1
Trajectory analysis for all cell clusters.
Detailed methods and data report of the developmental trajectory analysis for all cell clusters (15 clusters).
- https://cdn.elifesciences.org/articles/98096/elife-98096-fig3-data1-v2.pdf
-
Figure 3—source data 2
Cell counts per cluster by origin (control WNTi or treated WNTi + FGF8 organoids).
Summary datasheet showing WNTi or WNTi + FGF8 cell counts per cluster, extracted via 10 X Genomics Loupe Browser.
- https://cdn.elifesciences.org/articles/98096/elife-98096-fig3-data2-v2.xlsx
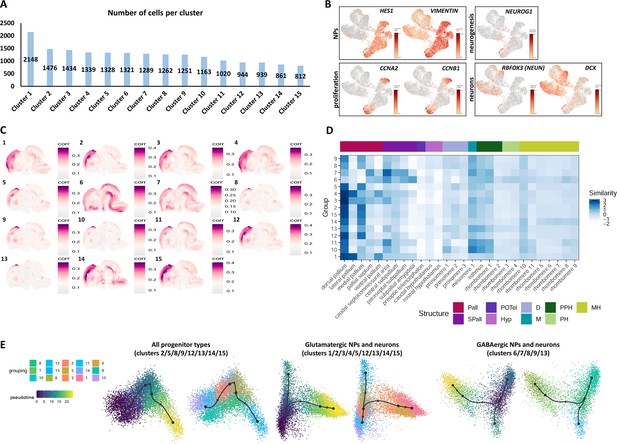
Single-cell RNA sequencing (scRNAseq) and VoxHunt similarity map analysis of 2-month-old telencephalic organoids.
(A) Number of cells per cluster after scRNAseq of day69 organoids. (B) Expression level of known markers identifying neural progenitor cells (NPs; VIMENTIN and HES1), proliferating progenitors (CCNA2 and CCNB1), differentiating neurons (NEUROG1) and post-mitotic neurons (DCX and RBFOX3, also called NEUN). Additional markers are illustrated in Figure 3C. (C) VoxHunt similarity map showing similarity correlation index (white to violet) of each cluster to reference databases of mouse regional brain atlases. (D) VoxHunt heatmap of similarity score showing the similarity degree (blue color code) between the expression profile of clusters (lines) and distinct regions of the mouse embryonic brain (columns). Note the high similarity between clusters 1/2/3/4/5/12/14/15 and dorsal-anterior pallium (neocortex). Clusters 6 and 7, on the contrary, show high similarity to ventral subpallial regions (ganglionic eminences). Legend on the right shows the brain area associated to each color. Pall: pallium; Spall: Subpallium; POTel: Preoptic telencephalon; Hyp: hypothalamus; D: diencephalon; M: mesencephalon; PPH: Prepontine hindbrain; PH: Pontine hindbrain; MH: Medullary hindbrain. (E) Trajectory analysis evaluating the most probable developmental trajectory linking all cell progenitor clusters (left), glutamatergic NPs and neurons (center) or GABAergic NPs and neurons (right). NP clusters can either convert into one another (suggesting plasticity to switch between dorsal and ventral telencephalic identity), either differentiate into post-mitotic neurons. In all cases, progenitors can also enter in a developmental ‘bottleneck’ represented by cluster 13 (CRYAB+ truncated aRGs).
-
Figure 3—figure supplement 1—source data 1
Cell count per cluster from scRNAseq analysis of day 69 organoids.
Summary datasheet showing the number of cells per cluster, extracted via 10 X Genomics Loupe Browser.
- https://cdn.elifesciences.org/articles/98096/elife-98096-fig3-figsupp1-data1-v2.xlsx
-
Figure 3—figure supplement 1—source data 2
VoxHunt analysis report.
Detailed report on the methods and data for VoxHunt visualization.
- https://cdn.elifesciences.org/articles/98096/elife-98096-fig3-figsupp1-data2-v2.pdf
-
Figure 3—figure supplement 1—source data 3
Trajectory analysis of progenitor cell clusters.
Detailed methods and data report on the developmental trajectory analysis for progenitor clusters (2, 5, 8, 9, 12, 13, 14, and 15).
- https://cdn.elifesciences.org/articles/98096/elife-98096-fig3-figsupp1-data3-v2.pdf
-
Figure 3—figure supplement 1—source data 4
Trajectory analysis of glutamatergic clusters.
Detailed methods and data report for the trajectory analysis of glutamatergic progenitors and neurons (clusters 1, 2, 3, 4, 5, 12, 13, 14, and 15).
- https://cdn.elifesciences.org/articles/98096/elife-98096-fig3-figsupp1-data4-v2.pdf
-
Figure 3—figure supplement 1—source data 5
Trajectory analysis of GABAergic clusters.
Detailed methods and data report for the trajectory analysis of GABAergic progenitors and neurons (clusters 6, 7, 8, 9, and 13).
- https://cdn.elifesciences.org/articles/98096/elife-98096-fig3-figsupp1-data5-v2.pdf
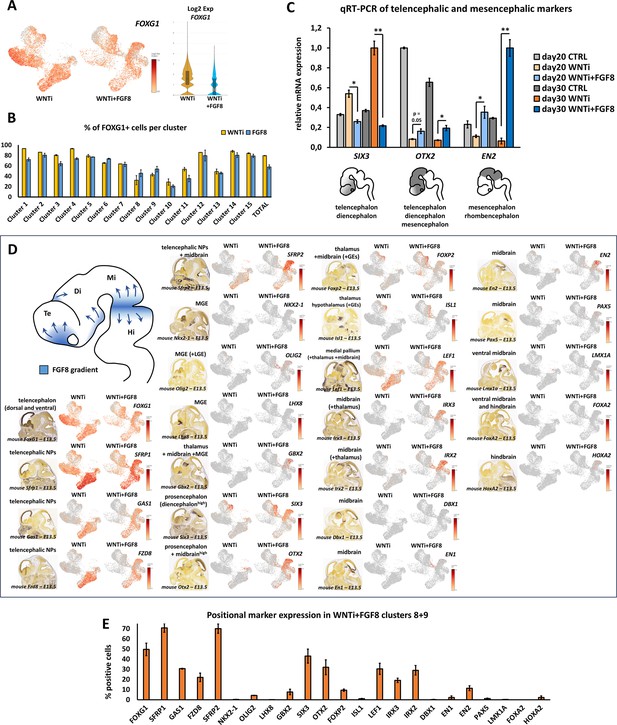
FGF8-dependent induction of diencephalic and mesencephalic markers in telencephalic organoids.
(A, B) Expression level and percentages of positive cells per cluster for FOXG1 in control (WNTi) and treated (WNTi + FGF8) organoids, as indicated. Despite a partial reduction of FOXG1 average expression (A), most of the cells in different clusters still express FOXG1 after FGF8 treatment (B). (C) Real time qRT-PCR quantification of telencephalic/diencephalic (SIX3), prosencephalic/midbrain (OTX2) and midbrain/hindbrain (EN2) markers in day20 or day30 control (CTRL), WNT inhibited (WNTi) or FGF8-treated (WNTi + FGF8) organoids, as indicated. FGF8 induces loss of SIX3 expression and increase of diencephalon/midbrain markers (OTX2, EN2), suggesting that long term FGF8 treatment can induce additional -more posterior- regional identities together with FOXG1+ cells. n=3 organoids per condition, pooled before RNA extraction. (D) At the top left of the panel, an early fetal brain scheme shows FGF8 sources (blue) in the anterior telencephalon, in the diencephalon and at the midbrain/hindbrain border, and their presumable diffusing gradients (arrows). The panel illustrates expression levels of distinct brain regional markers in UMAP projections of WNTi or WNTi + FGF8 single cell RNA sequencing samples, as indicated. Mouse brain images at embryonic day (E) 13.5 obtained from the Allen Brain Atlas show the main domains where these markers are found. (E) Graph showing the percentage of cells expressing different markers (indicated on x-axis) in clusters 8 and 9 of WNTi + FGF8 organoids. Clusters 8/9 indicate high percentage of cells expressing telencephalic (FOXG1, SFRP1, GAS1, SFRP2) and diencephalic/midbrain genes (SIX3, OTX2, LEF1, IRX3, IRX2, EN2). Very few or no cells express markers for the medial ganglionic eminence (GBX2, LHX8, NKX2-1, OLIG2), for the ventral midbrain (FOXP2, ISL1, LMX1A, FOXA2) or for more posterior regions (HOXA2).
-
Figure 3—figure supplement 2—source data 1
Number of FOXG1+ cells per cluster.
Summary datasheet showing the number of FOXG1+ cells in each cluster, extracted via 10 X Genomics Loupe Browser.
- https://cdn.elifesciences.org/articles/98096/elife-98096-fig3-figsupp2-data1-v2.xlsx
-
Figure 3—figure supplement 2—source data 2
Quantitative RT-PCR data for telencephalic and mesencephalic markers in human organoids.
Raw data of SIX3, OTX2, and EN2 expression in day 20 and day 30 CTRL, WNTi, and WNTi + FGF8 organoids.
- https://cdn.elifesciences.org/articles/98096/elife-98096-fig3-figsupp2-data2-v2.xls
-
Figure 3—figure supplement 2—source data 3
Number of cells expressing distinct positional markers in clusters 8 and 9.
Summary datasheet of cells from clusters 8 and 9 expressing different positional identity markers, extracted via 10 X Genomics Loupe Browser.
- https://cdn.elifesciences.org/articles/98096/elife-98096-fig3-figsupp2-data3-v2.xlsx
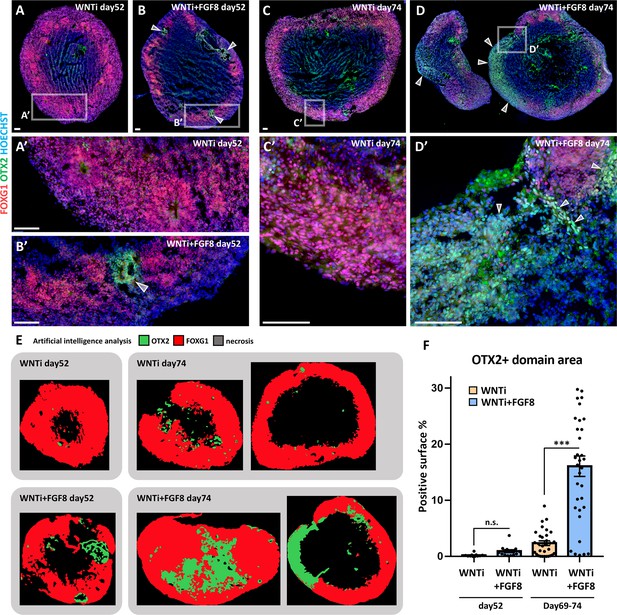
FGF8-dependent induction of OTX2+ domains in multi-regional organoids.
(A-D’) FOXG1 (red) and OTX2 (green) immunostaining in control (WNTi; A, A’, C, C’) and treated (WNTi + FGF8; B, B’, D, D’) organoids, showing the presence of FOXG1+ telencephalic domains and of OTX2+ FOXG1- diencephalic/mesencephalic domains at day52 (A-B’) and at day74 (C-D’). White arrowheads point to FOXG1- OTX2+ non-telencephalic areas in FGF8-treated organoids (high magnifications in B’ and D’). (E, F) Artificial intelligence analysis by HALO software of marker distribution in day52 and in day74 control or treated organoids, as indicated. Exemplificative images of automatically detected OTX2+ (green), FOXG1+ (red) and necrotic (black) regions are illustrated. Graph in F shows the percentages of OTX2+ surfaces in organoid sections, at different time points as indicated (quantified by HALO software; n≥8 sections from n≥3 organoids from n≥1 batch for each time point).
-
Figure 3—figure supplement 3—source data 1
OTX2+ area percentages in multi-regional organoids.
GraphPad sheet with OTX2+ area percentages in WNTi and WNTi + FGF8 organoid cryostat slices from day 52 to day 74 following OTX2 immunostaining.
- https://cdn.elifesciences.org/articles/98096/elife-98096-fig3-figsupp3-data1-v2.zip
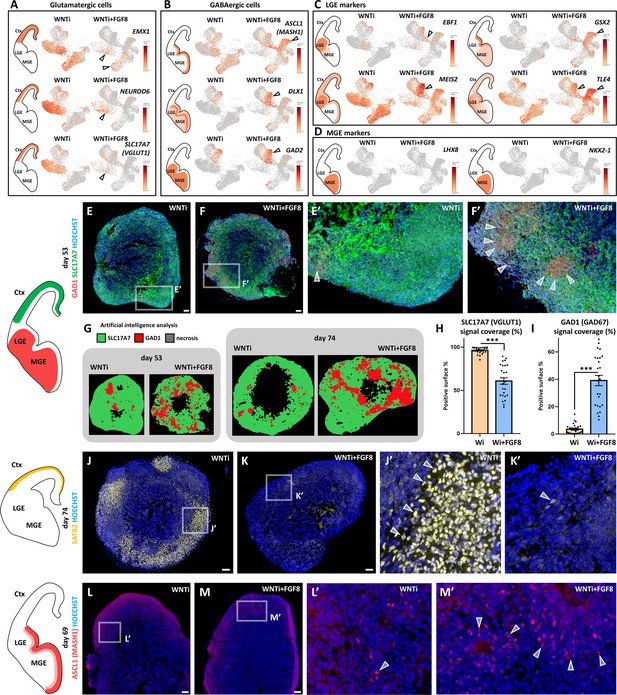
Effects of FGF8 treatment on the dorso-ventral cellular composition of telencephalic organoids.
(A, B) Expression levels of markers identifying dorsal glutamatergic NPs and neurons (Panel A; EMX1, NEUROD6, SLC17A7) and ventral GABAergic NPs and neurons (Panel B; ASCL1, DLX1, GAD2). Black arrowheads highlight clusters with the most notable changes in marker and cell abundance after FGF8 treatment. (C,D) Expression levels of known markers identifying ventral GABAergic NPs and neurons in the lateral ganglionic eminence (LGE) (Panel C; EBF1, GSX2, MEIS2 and TLE4) and the medial ganglionic eminence (MGE) (Panel D; NKX2-1 and LHX8). Black arrowheads in C highlight clusters with the largest differences in LGE marker and cell abundance following FGF8 treatment. (E-F’) GAD1 (red) and SLC17A7 (green) immunostaining in day53 control (WNTi) and treated (WNTi + FGF8) organoids, as indicated. The distribution of these markers in vivo is shown in the brain scheme on the left. (G–I) HALO software artificial intelligence (AI) analysis of marker distribution in day53 and day74 organoids. Representative images display areas automatically identified as SLC17A7+ (green), GAD1+ (red), and necrotic (black). Graphs in H and I show the proportions of SLC17A7+ (H) and GAD1+ (I) surface areas in day 74 organoid sections, as quantified by HALO AI; n≥8 sections from n≥4 organoids from n=1 batch per time point. (J-K’) Immunostaining for SATB2 (yellow) in day74 WNTi and WNTi + FGF8 organoids, as indicated. The left schematic depicts the in vivo distribution of SATB2+ neurons, and Figure 4—figure supplement 2 quantifies SATB2+ neuron density in these and additional samples. (L-M’) Immunostaining for ASCL1 (red) in day69 WNTi and WNTi + FGF8 organoids, as indicated. Left schematic shows the in vivo distribution of ASCL1+ ventral progenitors, with cell density detailed in Figure 4—figure supplement 2. Scale bars: 100 µm. Ctx, cortex; MGE, medial ganglionic eminence; LGE, lateral ganglionic eminence.
-
Figure 4—source data 1
SLC17A7 (VGLUT1)-positive area in human organoids.
GraphPad sheet with SLC17A7-positive area data in WNTi and WNTi + FGF8 organoids.
- https://cdn.elifesciences.org/articles/98096/elife-98096-fig4-data1-v2.zip
-
Figure 4—source data 2
GAD1 (GAD67)-positive area in human organoids.
GraphPad sheet with GAD1-positive area data in WNTi and WNTi + FGF8 organoids.
- https://cdn.elifesciences.org/articles/98096/elife-98096-fig4-data2-v2.zip
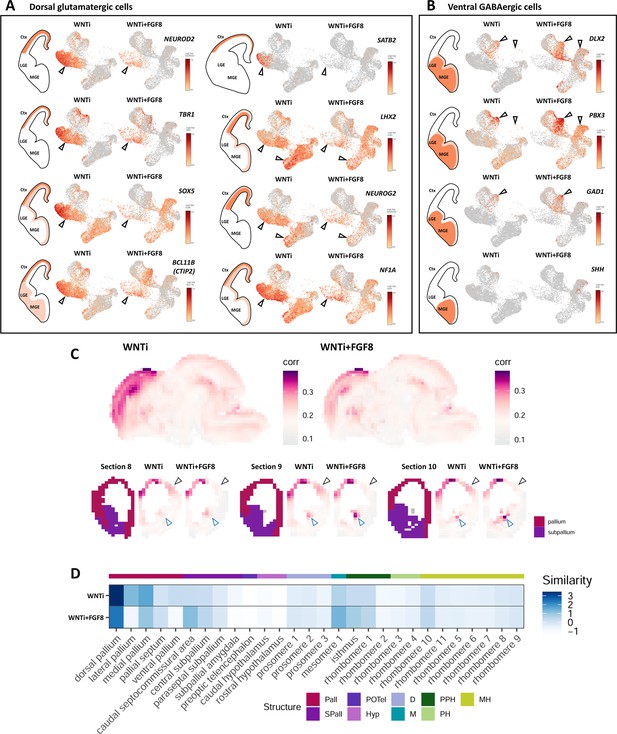
Changes in cellular composition and glutamatergic/GABAergic identity upon FGF8 treatment.
(A,B) Expression level of known markers identifying dorsal glutamatergic NPs and neurons (panel A; NEUROD2, TBR1, SOX5, BCL11B -also called CTIP2-, SATB2, LHX2, NEUROG2 and NF1A) or ventral GABAergic NPs and neurons (panel B; DLX2, PBX3, SHH and GAD1). FGF8 treatment causes an increase of ventral GABAergic cells at the expense of dorsal glutamatergic ones. Empty arrowheads indicate clusters showing the greatest differences in cell and marker abundance following FGF8 treatment. (C) VoxHunt similarity map showing correlation index (white to violet color code) of WNTi (left) or WNTi + FGF8 (right) organoids to reference databases of mouse regional brain atlases. For this analysis, all WNTi cells and all WNTi + FGF8 cells are evaluated. FGF8-treated organoids show lower similarity to dorso-lateral pallium (black arrowheads) and increased similarity to subpallial domains (blue arrowheads), compared to control samples. (D) VoxHunt heatmap illustrating the similarity score (blue color code) among the expression profile of organoid samples (WNTi -upper line- and WNTi + FGF8 -lower line-) and distinct mouse brain regions (columns). FGF8-treated organoids show lower similarity to dorsal pallium and increased similarity to subpallial regions, compared to control samples. Pall: pallium; Spall: Subpallium; POTel: Preoptic telencephalon; Hyp: hypothalamus; D: diencephalon; M: mesencephalon; PPH: Prepontine hindbrain; PH: Pontine hindbrain; MH: Medullary hindbrain.
-
Figure 4—figure supplement 1—source data 1
VoxHunt analysis report.
Detailed report on VoxHunt visualization methods and data.
- https://cdn.elifesciences.org/articles/98096/elife-98096-fig4-figsupp1-data1-v2.pdf
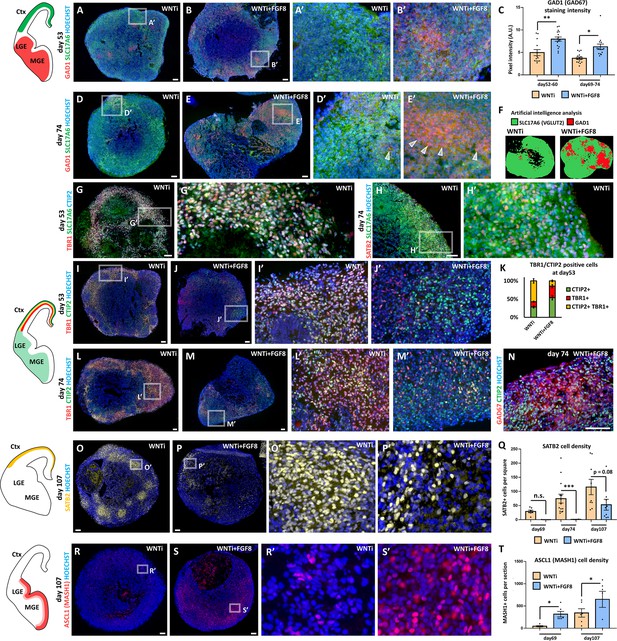
Immunostaining analysis of glutamatergic and GABAergic cellular composition of control and FGF8-treated telencephalic organoids.
(A–F) GAD1 (also called GAD67; red) and SLC17A6 (also called VGLUT2; green) immunostaining in day53 and in day74 control (WNTi) and treated (WNTi + FGF8) organoids, as indicated. The distribution of these markers in vivo is illustrated in the brain scheme on the left. Graph in C displays GAD1 pixel intensity quantification at day52-60 and at day69-74, while F shows exemplificative images of automatically detected SLC17A6 (green) and GAD1 (red) marker distribution in day74 organoids (HALO software). n≥13 sections from n≥4 organoids from n=2 batches. (G-H’) Co-staining of SLC17A6 (green) with TBR1 and CTIP2 (red and blue in G,G’) and SATB2 (red in H,H’), highlighting co-expression of SLC17A6 with neocortical markers. (I-M’) TBR1 (red) and CTIP2 (green) immunostaining in day53 (I–J) or day74 (L-M’) control (WNTi) and treated (WNTi + FGF8) organoids, as indicated. The distribution of these markers in vivo is depicted in the brain schemes on the left. Note that TBR1 and CTIP2 can be co-expressed dorsally (in cortical layers) during development; TBR1 is exclusively dorsal, whereas CTIP2 is also expressed in the ventral telencephalon. Graph in K shows the percentage of TBR1+ (red), CTIP2+ (green) or double TBR1+/CTIP2+ (yellow) cells in WNTi or WNTi + FGF8 organoids at different steps (day53 in H and day74 in I), as indicated. (N) GAD67 (red) and CTIP2 (green) immunostaining in day74 WNTi + FGF8 organoids with double CTIP2+ GAD67+ cells. (O–Q) SATB2 (yellow) immunostaining in day107 control (WNTi) and treated (WNTi + FGF8) organoids, as indicated. The distribution of SATB2+ neurons in vivo is illustrated in the brain scheme on the left, while graph in Q displays cell density at day69, day74 or day107. SATB2 cell density was measured as number of SATB2+ cells per 150X150 µm square; n≥4 sections from n≥2 organoids from n=1 batch per time point/condition. (R–T) ASCL1 (also known as MASH1; red) immunostaining in day107 control (WNTi) and treated (WNTi + FGF8) organoids, as indicated. Quantification of the cell density (number of cells per organoid section) of ASCL1+ progenitors at day69 or day107 is shown in the graph in T; n≥5 sections from n≥2 organoids from n=1 batch per time point/condition. Scale bars: 100 µm.
-
Figure 4—figure supplement 2—source data 1
GAD1 (GAD67) pixel intensity in human organoids.
GraphPad sheet with raw pixel intensity data measured in WNTi and WNTi + FGF8 organoid cryostat slices following GAD1 immunostaining.
- https://cdn.elifesciences.org/articles/98096/elife-98096-fig4-figsupp2-data1-v2.zip
-
Figure 4—figure supplement 2—source data 2
TBR1 and CTIP2 co-expression in human organoids.
Excel summary datasheet showing TBR1 and CTIP2 cell percentages in WNTi and WNTi + FGF8 organoids.
- https://cdn.elifesciences.org/articles/98096/elife-98096-fig4-figsupp2-data2-v2.xlsx
-
Figure 4—figure supplement 2—source data 3
SATB2 cell abundance in human organoids.
GraphPad sheet with numbers of SATB2+ cells in WNTi and WNTi + FGF8 organoid cryostat slices.
- https://cdn.elifesciences.org/articles/98096/elife-98096-fig4-figsupp2-data3-v2.zip
-
Figure 4—figure supplement 2—source data 4
ASCL1 (MASH1) cell abundance in human organoids.
GraphPad sheet with ASCL1+ cell counts in WNTi and WNTi + FGF8 organoid cryostat slices.
- https://cdn.elifesciences.org/articles/98096/elife-98096-fig4-figsupp2-data4-v2.xlsx
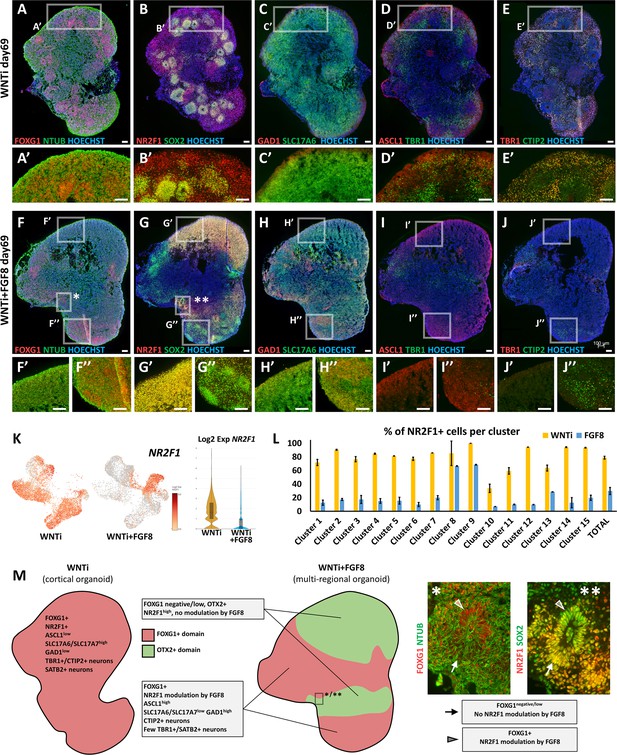
Regional identity-dependent effect of FGF8 treatment on D/V identity and target gene modulation.
(A-E’) Immunostaining of key neural and regional markers on day69 WNTi control organoids; FOXG1 (red in A,A’), NR2F1 (red in B,B’) and SOX2+ NPs (green in B,B’). Glutamatergic/cortical markers SLC17A6 (also called VGLUT2; green in C,C’), TBR1 (green in D,D’ or red in E,E’) and CTIP2 (green in E,E’) are abundant, indicating neocortical identity. (F-J’’) Immunostaining of the same key neural and regional markers showed in A-E’, but on sections of day69 WNTi + FGF8 treated organoids. Insets represent magnification from a FOXG1-negative non-telencephalic region (F’, G’, H’, I’, J’) or from a FOXG1+ telencephalic region (F’’, G’’, H’’, I’’, J’’). NR2F1 (red in G-G’’) show high intensity, i.e. no response to FGF8 treatment, in FOXG1-negative non-telencephalic areas. Only FOXG1+ telencephalic domains exhibit FGF8-mediated reduction of NR2F1 levels (G, G’’), associated with induction of ventral markers GAD1 (red in H,H’’) and ASCL1 (red in I,I’’) at the expense of dorsal cortical markers TBR1 (green in I-I’’). No dorsal cortical markers such as TBR1 and CTIP2 are detectable in FOXG1-negative regions (I’, J’), illustrating that efficient acquisition of anterior identity is necessary for production of cortical neurons in organoids. Insets indicated by asterisks in F (*) and G (**) are showed at high magnification in M. Scale bars: 100 µm. (K,L) Expression level and percentages of positive cells per cluster for NR2F1 in control (WNTi) and treated (WNTi + FGF8) organoids, as indicated. NR2F1 transcript levels are greatly reduced by FGF8 (K); however, clusters 8 and 9 fail to respond to FGF8 treatment by downregulating NR2F1 (L) and still contain >60% NR2F1+ cells. (M) Schematic representation of distinct domains in control (WNTi; left) and FGF8-treated (WNTi + FGF8; right) organoids. FOXG1+ telencephalic WNTi organoids (left) have high levels of the glutamatergic marker SLC17A6 and abundant SATB2+ and TBR1/CTIP2 double positive cortical neurons. Upon FGF8 treatment, WNTi + FGF8 organoids (right) develop co-existing FOXG1+ telencephalic domains (light red areas), in which ventral GABAergic markers GAD1 and ASCL1 are upregulated at the expense of dorsal genes, and FOXG1-/OTX2+ diencephalic/midbrain domains (light green areas). Immunostaining magnifications on the right (magnification * and ** from images in F and G, respectively) show a neural rosette located at the border between a FOXG1+ domain (upper half of the rosette; arrowheads) and an adjacent FOXG1- low expressing domain (lower half; arrows). This highlights that NR2F1 is down-regulated by FGF8 only in the telencephalic SOX2+ domain (upper half of the rosette), indicating that FGF8 target genes can respond in a different way depending on the regional identity of organoid domains. Scale bars: 100 µm.
-
Figure 4—figure supplement 3—source data 1
Percentage of NR2F1+ cells per cluster in WNTi and WNTi + FGF8 organoids.
Summary Excel datasheet showing NR2F1-positive cell counts per cluster for WNTi and WNTi + FGF8 samples, extracted via 10 X Genomics Loupe Browser.
- https://cdn.elifesciences.org/articles/98096/elife-98096-fig4-figsupp3-data1-v2.xlsx
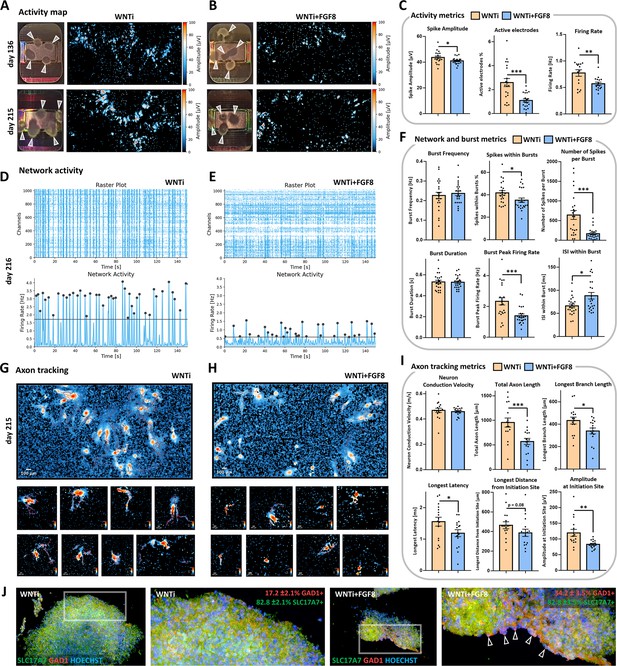
Electrophysiological spontaneous activity of control and FGF8-treated 3D organoid networks.
(A–C) Spontaneous activity maps showing mean spike amplitude in 4-month-old (day136; upper row) and 7-month-old (day215; lower row) control (WNTi; A) and treated organoids (WNTi + FGF8; B), 2–3 weeks post-plating on high-density MEA chips. Mean activity metrics from three independent batches are summarized in (C), showing a general reduction in spike amplitude, percentage of active electrodes, and firing rate in FGF8-treated organoids. White arrowheads in electrode images indicate organoids fully or partially adhered to the MEA recording surface; note that high activity levels are observed at the organoid edges, where axonal tracts extend. (D–F) Temporal raster plots displaying firing events recorded by the 1024 most active electrodes (upper graphs) and their synchronicity indicative of network activity (lower graphs) in 7-month-old controls (WNTi; D) and FGF8-treated organoids (WNTi + FGF8; E), 3 weeks post-plating. Note that the instrument automatically sets the detection threshold (black bar) at varying levels, based on the average baseline activity specific to each sample (see Materials and methods). Graphs in (F) show network metrics (Burst frequency; Spikes within bursts) and burst metrics (number of spike per burst, burst peak firing rate, burst duration, inter-spike interval within bursts). (G–I) Overview fields of axon tracts (upper row) and representative images of individual neuronal tracts (lower rows) extending from organoids on the MEA surface, as detected using the automatic axon tracking function in WNTi (G) and WNTi + FGF8 (H) samples at day215. Graphs in (I) present axon tracking metrics (network conduction velocity, total axon or branch lengths, longest signal latency, maximum distance from initiation site, and signal amplitude), showing reduced signal amplitude and spatial propagation in FGF8-treated organoids. Additional axon tract images at different stages of differentiation are provided in Figure 5—figure supplement 1. (J) Immunostaining for GAD1 (red) and SLC17A6 (green) in day 145 control (WNTi) and treated (WNTi + FGF8) organoids, following detachment from MEA chips post-recording. Percentages of GAD1- and SLC17A7-positive tissue in organoids are indicated. For all graphs: data represent n=3 distinct batches (each with 2–4 organoids on the MEA chip, see Materials and methods).
-
Figure 5—source data 1
Analysis of spontaneous network activity in WNTi and WNTi + FGF8 organoids on MEAs.
Excel summary datasheet containing raw activity scan, network analysis, and axon tracking tool metrics for WNTi and WNTi + FGF8 organoids on MEA electrodes.
- https://cdn.elifesciences.org/articles/98096/elife-98096-fig5-data1-v2.xlsx
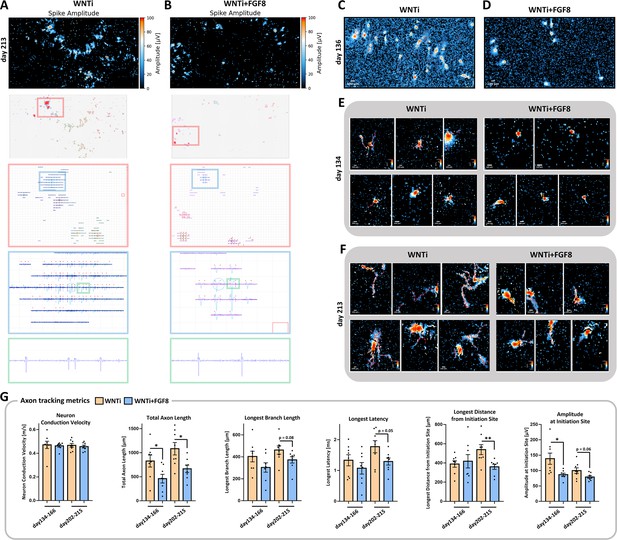
Representative spike traces and axonal maturation of WNTi and WNTi + FGF8 organoids on HD-MEA.
(A, B) Footprints of action potentials in control WNTi (A; left column) and treated WNTi + FGF8 (B; right column) organoids, as automatically detected by MaxOne software. Progressive higher magnifications of propagating spikes are displayed in red and blue boxes, while representative traces displaying the shape of action potentials are shown in the bottom row (green boxes). (C, D) Overview fields of axon tracts extending from organoids on MEA surface, as detected by the automatic axon tracking function, in WNTi (C) and WNTi + FGF8 (D) samples at day136. (E, F) Exemplificative images of single neuronal tracts extending from organoids on MEA surface, in WNTi (left) and WNTi + FGF8 (right) samples at day134 (E) and at day213 (F). Note the progressive increase in complexity and length of axons, suggesting maturation of the organoids, which is more limited after treatment with FGF8. (G) Graphs show axon tracking metrics (network conduction velocity, total axon or total branch lengths, longest signal latency, longest distance from and signal amplitude at the initiation site), in control (orange columns) and in FGF8-treated organoids (blue columns) at different developmental stages, as indicated. For graphs in G: data from n=2 distinct batches at two different differentiation times (each batch including 2–4 organoids on the MEA chip, see Materials and methods).
-
Figure 5—figure supplement 1—source data 1
Axon tracking metrics in WNTi and WNTi + FGF8 organoids on MEAs.
Excel summary datasheet with raw axon tracking measurements for control WNTi and FGF8-treated organoids on MEAs.
- https://cdn.elifesciences.org/articles/98096/elife-98096-fig5-figsupp1-data1-v2.xlsx
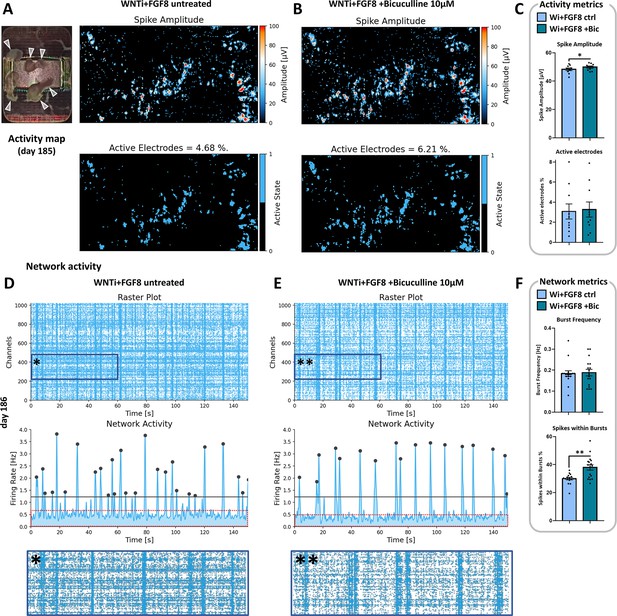
Bicuculline-mediated inhibition of GABA-A receptors and its effect on WNTi + FGF8 organoid spontaneous activity.
(A–C) Spontaneous activity maps showing the mean spike amplitude of 6-month-old WNTi + FGF8 organoids, before (left) or after (right) treatment with 10 µM Bicuculline, a chemical inhibitor of GABA-A receptor. Mean values of activity metrics are shown in C and highlight a slight but significant increase of spike amplitudes. White arrowheads in the electrode image on the top left point to organoids adhering totally or partially on the MEA recording surface. (D–F) Temporal raster plots showing firing events recorded by the 1024 most active electrodes (upper graphs) and their synchronicity suggestive of network activity (lower graphs), as detected from untreated (left) or Bicuculline-treated (right) WNTi + FGF8 organoids. Note that the background noise due to random activity falling outside of synchronous bursts (red dotted boxes) is decreased upon treatment with Bicuculline, as also visible in high magnification insets (* and **; before and after Bicuculline treatment, respectively). Graphs in F depict network metrics (Burst frequency and Spikes within bursts), highlighting a significant increase of percentage of spikes within synchronous network bursts. For all graphs: data from n=2 distinct batches (each including 2–4 organoids on the MEA chip, see Materials and methods).
-
Figure 5—figure supplement 2—source data 1
Activity scan and network analysis of WNTi + FGF8 organoids on MEAs with or without Bicuculline treatment.
Excel summary datasheet with raw activity and network metrics for WNTi + FGF8 organoids before and during Bicuculline treatment.
- https://cdn.elifesciences.org/articles/98096/elife-98096-fig5-figsupp2-data1-v2.xlsx
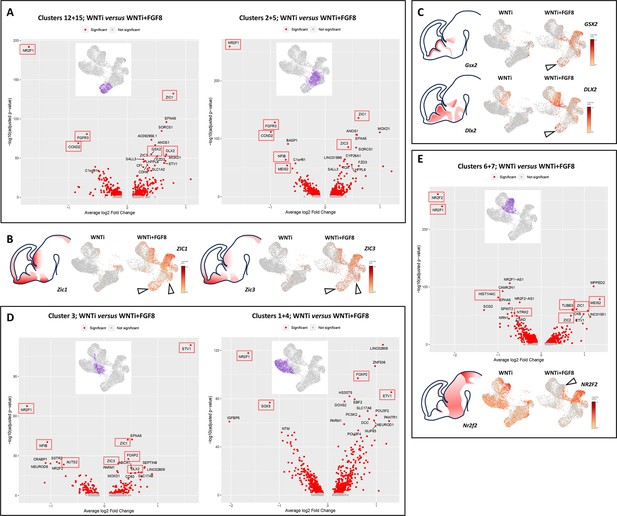
Analysis of differentially expressed genes (DEGs) in glutamatergic and GABAergic progenitors and neurons upon FGF8 treatment.
(A) DEG analysis comparing control (WNTi) and treated (WNTi + FGF8) organoids, highlighting the most strongly (x-axis, average log2 fold change) and most significantly (y-axis, adjusted p-value in -log10) differentially expressed genes (DEGs) in clusters 12/15 (proliferating glutamatergic progenitors; left volcano plot) or clusters 2/5 (non-proliferating glutamatergic progenitors; right volcano plot). Red dots indicate significantly (adjusted P-value <0.05) regulated genes, with the names of the 20 most significant ones displayed (see source data material for a complete list of DEGs). (B) Expression level of ZIC1 (left) and ZIC3 (right) in UMAP projections of WNTi and WNTi + FGF8 samples, as indicated. Black arrowheads point to increased ZIC1 and ZIC3 expression in glutamatergic progenitor clusters following FGF8 treatment. (C) Expression level of GSX2 (upper panel) and DLX2 (lower panel) in UMAP projections of WNTi or WNTi + FGF8 samples, as indicated. Black arrowheads indicate increased GSX2 and DLX2 expression in proliferating glutamatergic progenitors upon FGF8 treatment. (D) DEG analysis comparing control (WNTi) and treated (WNTi + FGF8) organoids, highlighting the most significantly regulated genes in cluster 3 (early differentiating glutamatergic neurons; left volcano plot) or clusters 1/4 (differentiated glutamatergic neurons; right volcano plot). (E) DEG analysis comparing control (WNTi) and treated (WNTi + FGF8) organoids, showing the most significantly regulated genes in clusters 6/7 (volcano plot), corresponding to GABAergic neurons. The panel below shows the expression level of NR2F2 in UMAP projections of WNTi and WNTi + FGF8 samples, as indicated. The black arrowhead points to decreased NR2F2 expression in GABAergic cells following FGF8 treatment. Red boxes highlight FGF target genes or genes noted in OMIM as disease-related. Brain schematics with gene expression patterns are based on embryonic day 13.5 stainings from the Mouse Allen Brain Atlas.
-
Figure 6—source data 1
Analysis of differentially expressed genes in cycling glutamatergic progenitors following FGF8 treatment.
Excel file listing genes with differential expression between WNTi and WNTi + FGF8 organoids in clusters 12 and 15 (glutamatergic cycling progenitors), visualized as a volcano plot in Figure 6A.
- https://cdn.elifesciences.org/articles/98096/elife-98096-fig6-data1-v2.xlsx
-
Figure 6—source data 2
Analysis of differentially expressed genes in non-cycling glutamatergic progenitors following FGF8 treatment.
Excel file listing genes with differential expression between WNTi and WNTi + FGF8 organoids in clusters 2 and 5 (glutamatergic non-cycling progenitors), visualized as a volcano plot in Figure 6A.
- https://cdn.elifesciences.org/articles/98096/elife-98096-fig6-data2-v2.xlsx
-
Figure 6—source data 3
Analysis of differentially expressed genes in early differentiating glutamatergic neurons following FGF8 treatment.
Excel file listing genes with differential expression between WNTi and WNTi + FGF8 organoids in cluster 3 (glutamatergic early differentiating neurons), visualized as a volcano plot in Figure 6D.
- https://cdn.elifesciences.org/articles/98096/elife-98096-fig6-data3-v2.xlsx
-
Figure 6—source data 4
Analysis of differentially expressed genes in post-mitotic glutamatergic neurons following FGF8 treatment.
Excel file listing genes with differential expression between WNTi and WNTi + FGF8 organoids in clusters 1 and 4 (glutamatergic neurons), visualized as a volcano plot in Figure 6D.
- https://cdn.elifesciences.org/articles/98096/elife-98096-fig6-data4-v2.xlsx
-
Figure 6—source data 5
Analysis of differentially expressed genes in GABAergic cells following FGF8 treatment.
Excel file listing genes with differential expression between WNTi and WNTi + FGF8 organoids in clusters 6 and 7 (GABAergic neurons), visualized as a volcano plot in Figure 6E.
- https://cdn.elifesciences.org/articles/98096/elife-98096-fig6-data5-v2.xlsx
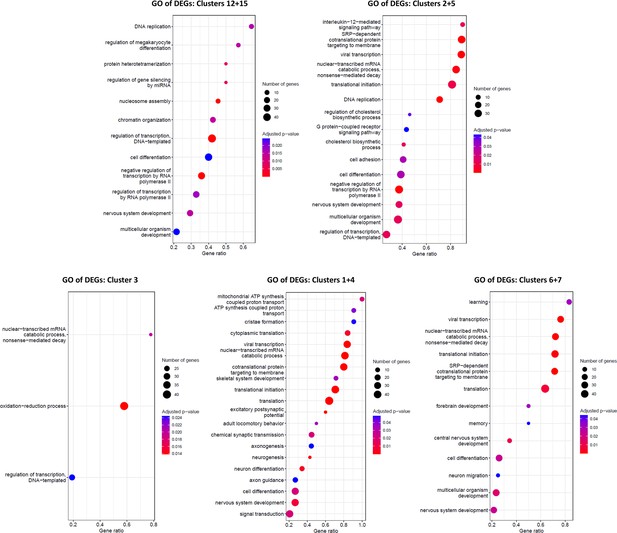
Enrichment analysis of Gene Ontology (GO) terms in distinct NP or neuronal clusters.
Results of enrichment analysis of the biological process GO terms performed with the gene set enrichment analysis (GSEA) method. Each graph shows the results relative to a specific cluster or subset of clusters, as indicated. Red to blue color code corresponds to adjusted p-value, while the circle size indicates the number of genes that belong to a specific GO term listed on the left of each graph.
-
Figure 6—figure supplement 1—source data 1
Gene Ontology (GO) enrichment analysis in NP and neuronal clusters.
Detailed report of enrichment analysis comparing different cell clusters in WNTi and WNTi + FGF8 organoids.
- https://cdn.elifesciences.org/articles/98096/elife-98096-fig6-figsupp1-data1-v2.pdf
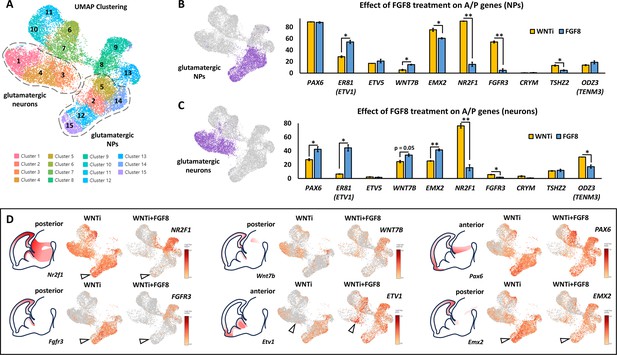
FGF8-dependent acquisition of distinct antero-posterior areal identities in human telencephalic organoids.
(A) Glutamatergic neural progenitor (NPs; clusters 2/5/12/14/15) and glutamatergic neurons (clusters 1/3/4) are highlighted on the UMAP projection of day69 organoid scRNAseq data. (B,C) Images on the right display the clusters selected for analysis, as described in A. The graphs show the percentages of cells expressing anterior-posterior (A/P) cortical markers within the two highlighted cluster groups: NPs (top graph) and neurons (bottom graph). The percentage of cells positive for anterior markers (PAX6, ER81, ETV5) and posterior markers (WNT7B, EMX2, NR2F1, FGFR3, CRYM, TSHZ2, ODZ3) is shown in yellow for control (WNTi) organoids and in blue for FGF8-treated (WNTi + FGF8) organoids. (D) Expression level of key posterior (NR2F1, FGFR3, WNT7B, EMX2) and anterior (ETV1, PAX6) genes in UMAP projections of WNTi or WNTi + FGF8 day69 organoid samples, as indicated. Black arrowheads in the NR2F1 and FGFR3 UMAP projections point to decreased expression in proliferating glutamatergic progenitors upon FGF8 treatment, while arrowheads in the ETV1 UMAP projection indicate increased expression in FGF8-treated glutamatergic neurons. Brain schematics with gene expression patterns are based on embryonic day 13.5 staining data from the Mouse Allen Brain Atlas.
-
Figure 7—source data 1
Percentage of cells expressing antero-posterior cortical markers in WNTi and WNTi + FGF8 organoids.
Summary Excel datasheet with counts of cells expressing A/P markers (PAX6, ER81, ETV5, WNT7B, EMX2, NR2F1, FGFR3, CRYM, TSHZ2, and ODZ3) within glutamatergic progenitor (clusters 2, 5, 12, 14, 15) and neuron (clusters 1, 3, 4) groups for both WNTi and WNTi + FGF8 samples, extracted via 10 X Genomics Loupe Browser.
- https://cdn.elifesciences.org/articles/98096/elife-98096-fig7-data1-v2.xlsx
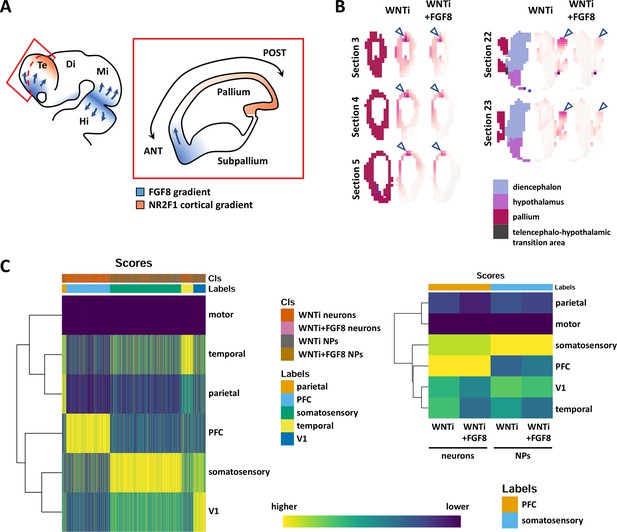
FGF8-dependent acquisition of different antero-posterior areal identities in human telencephalic organoids.
(A) Schematic drawing of the mammalian developing brain showing FGF8 sources (blue) in the anterior telencephalon and at the midbrain/hindbrain border, and their presumable diffusing gradients (arrows). The red inset illustrates a sagittal section of the telencephalon with the two opposite FGF8 (blue) and NR2F1 (orange) gradients. (B) VoxHunt Similarity Map showing similarity correlation index (white to violet color code) of control (WNTi) and treated (WNTi + FGF8) organoids on virtual brain coronal sections. The color code in the first left column of brain virtual sections identifies different brain regions, listed in the legend at the bottom. Blue arrowheads point to the dorsal-most region of the pallium. (C) Cell-level (left) and cluster-level (right) heatmaps of the SingleR assignment scores (i.e. the confidence of the predicted labels across the dataset; dark blue to yellow color code) as well as the corresponding inferred annotation for the clusters/cells in the ‘Labels’ top bar. Cell/cluster annotation was obtained by using SingleR to evaluate the similarity between control (WNTi) or FGF8-treated (WNTi + FGF8) samples against a 16 post conceptional week (PCW) fetal brain dataset. Glutamatergic progenitors correspond to clusters 2/5/12/14/15, while neurons corresponded to clusters 1/3/4. The reference dataset corresponds to primary cells dataset published by Speir et al., 2021 and where only cells belonging to 16 PCW and not to the hippocampus were kept. In the left graph, the ‘Cls’ top bar identifies the organoid cells. Note that most of the organoid cells (columns) are annotated as pre-fontal cortex (PFC; light blue) or somatosensory cortex (green), based on transcriptional similarity. In the graph on the right, the average annotation score per sample is depicted; control and treated progenitors show high annotation score to somatosensory cortex, while control and treated neurons resemble the PFC. Despite this, note that FGF8 treatment decreases the annotation score to caudal V1 (visual) and temporal areas, while slightly increasing the annotation score to the rostral PFC, indicating anteriorization of cell identity.
-
Figure 7—figure supplement 1—source data 1
SingleR cluster annotation analysis of human organoid areal identity.
Detailed report on methods and data for annotating clusters/cells using SingleR, comparing in vitro organoids with in vivo fetal cortical regions, specifically assessing glutamatergic neuron (clusters 1, 3, 4) and progenitor (clusters 2, 5, 12, 14, 15) clusters.
- https://cdn.elifesciences.org/articles/98096/elife-98096-fig7-figsupp1-data1-v2.pdf
Tables
| Reagent type (species) or resource | Designation | Source or reference | Identifiers | Additional information |
|---|---|---|---|---|
| Cell line (Homo sapiens) | HMGU1; Human induced pluripotent stem cells | kind gift of Dr. Drukker | HMGU1; source cells: BJ (ATCC CRL-2522) | MTA approval was obtained from the Helmholtz Zentrum München (HMGU), Germany |
| Peptide, recombinant protein | Recombinant Human/Mouse FGF-8b Protein | R&D | 423-F8-025/CF | (100 ng/ML) |
| Peptide, recombinant protein | BDNF | PeproTech | #450–02 | 10 ng/mL |
| Commercial assay or kit | hPSC genetic Analysis kit | Stem Cell Technologies | #07550 | hiPSCs culture |
| Commercial assay or kit | NucleoSpin RNA II columns | Macherey-Nagel | 740902.50 | hiPSCs culture |
| Commercial assay or kit | Reverse Transcriptase Core Kit | Eurogentec | RT-RTCK-03 | hiPSCs culture |
| Chemical compound, drug | Matrigel | Corning | 354234 | 5–10 µl matrigel dissolved in 1 ml cold DMEM-F12 for each well of a 6-well culture plate |
| Chemical compound, drug | mTeSR1 medium | STEMCELL Technologies | #85850 | hiPSCs culture |
| Chemical compound, drug | ROCK inhibitor Y-27632 | MedChemExpress; or Stem Cell Technologies | MedChemExpress HY-10583; or Stem Cell Technologies #72304 | 10 µM |
| Chemical compound, drug | DMEM/F12 | Thermo Fisher Scientific | #31331028 | hiPSCs culture |
| Chemical compound, drug | N-2 Supplement 100X | Thermo Fisher Scientific | 17502–048 | hiPSCs culture |
| Chemical compound, drug | B-27 Supplement 50X, minus vitamin A | Thermo Fisher Scientific | 12587–010 | hiPSCs culture |
| Chemical compound, drug | LDN-193189 | Sigma | SML0559-5MG | 0.25 µM |
| Chemical compound, drug | SB-431542 | Sigma | S4317-5MG | 5 µM |
| Chemical compound, drug | XAV-939 | Stem cell technologies | 72674 | 2 µM |
| Antibody | CTIP2, FOXG1, GAD1, OTX2, MAP2, ASCL1, NR2F1, NTUB (ACETILATED TUBULIN), OCT-3/4, PAX6, SATB2, SOX2, TBR1, TUJ1, SLC17A6, SLC17A7 antibodies | See Table 2 for supplier information and antibody host species | See Table 2 for reference codes | See Table 2 for antibody dilution |
| Sequence-based reagent | DUSP6, EN2, ETV1, ETV4, ETV5, FOXG1, GAPDH, NANOG, NR2F1, OCT4, OTX2, PAX6, SIX3, SOX2, SPRY4, B-ACTIN primers | This paper | PCR primers | See Table 1 for sequence of forward and reverse primers |
Primer sequences used.
| gene | forward sequence | reverse sequence |
|---|---|---|
| DUSP6 | AACCTGTCCCAGTTTTTCCCT | GCCAAGCAATGTACCAAGACAC |
| EN2 | TCTACTGTACGCGCTACTCG | CGCTTGTCCTCTTTGTTCGG |
| ETV1 | CCAATAGTCAGCGTGGGAGAA | TTCTGCAAGCCATGTTTCCTGT |
| ETV4 | TCAAACAGGAACAGACGGACTT | AGGTTTCTCATAGCCATAGCCCA |
| ETV5 | ACACGGGTTCCAGTCACCAA | GCTGCTGGAGAAATAACCCCC |
| FOXG1 | GCCAAGTTTTACGACGGGAC | AAGGGTTGGAAGAAGACCCC |
| GAPDH | CGTGGAAGGACTCATGACCA | CAGTCTTCTGGGTGGCAGTGA |
| NANOG | CAAAGGCAAACAACCCACTT | TCTGCTGGAGGCTGAGGTAT |
| NR2F1 | TGGCAATGGTAGTTAGCAGCT | TTGAGGCACTTCTTGAGGCG |
| OCT4 | GTGGAGGAAGCTGACAACAA | ATTCTCCAGGTTGCCTCTCA |
| OTX2 | CCTCACTCGCCACATCTACT | CTTGGTGGGTGGGTTTGGAG |
| PAX6 | AGTGAATCAGCTCGGTGGTGTCTT | TGCAGAATTCGGGAAATGTCGCAC |
| SIX3 | CAGCAAGAAACGCGAACTGG | AATGGCCTGGTGCTGGA |
| SOX2 | AGGATAAGTACACGCTGCCC | TAACTGTCCATGCGCTGGTT |
| SPRY4 | CGGCTTCAGGATTTACACAGAC | CTGCAAACCGCTCAATACAGG |
| β-Actin | CTTCGCGGGCGACGAT | ACATAGGAATCCTCCTGACCC |
Primary antibodies used.
| Primary antibody | species | Concentration used | Brand | ref |
|---|---|---|---|---|
| CTIP2 | Rat monoclonal | (1:1000) | Abcam | ab18465 |
| FOXG1 | Rabbit polyclonal | (1:1000) | Abcam | ab18259 |
| GAD1 | Mouse monoclonal | (1:1000) | Millipore | MAB5406 |
| OTX2 | Goat polyclonal | (1:1000) | R&D | AF1979 |
| MAP2 | Mouse monoclonal | (1:1000) | Sigma | M4403 |
| ASCL1 (MASH1) | Mouse monoclonal | (1:1000) | BD Biosciences | 556604 |
| NR2F1 (COUP TF1) | Rabbit monoclonal | (1:1000) | Abcam | ab181137 |
| NTUB | Mouse monoclonal | (1:1000) | Sigma | T6793 |
| OCT-3/4 | Mouse monoclonal | (1:500) | Santa Cruz | sc-5279 |
| PAX6 | Rabbit polyclonal | (1:1000) | Millipore | AB2237 |
| SATB2 | Mouse monoclonal | (1:1000) | Abcam | ab51502 |
| SOX2 | Mouse monoclonal | (1:1000) | R&D | MAB2018 |
| TBR1 | Rabbit polyclonal | (1:1000) | Abcam | ab31940 |
| TUJ1 (TUBB3) | Rabbit polyclonal | (1:1000) | BioLegend | 802001 |
| SLC17A6 (VGLUT2) | Guinea Pig polyclonal | (1:1000) | Millipore | AB2251 |
| SLC17A7 | Rabbit polyclonal | (1:1000) | Invitrogen | 48–2400 |