A molecular proximity sensor based on an engineered, dual-component guide RNA
Figures
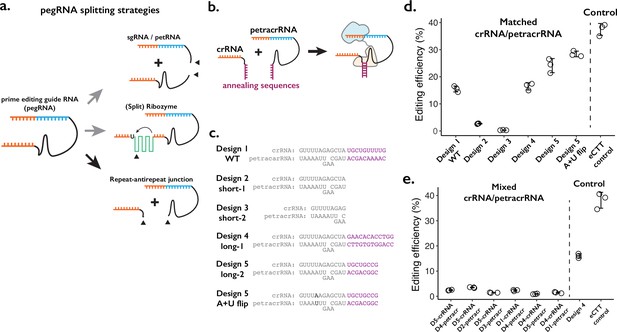
Testing the consequences of splitting the prime editing guide RNA (pegRNA) at the repeat:anti-repeat junction.
(a) We tested three classes of split-pegRNA designs: (Top) pegRNA is split into sgRNA and petRNA. (Center) Self-splicing ribozyme is inserted into pegRNA, thus splitting it into two parts. (Bottom) The Cas9-binding scaffold is split at the crRNA:tracrRNA junction, which is joined through a GAAA RNA tetraloop in the standard sgRNA. (b) Dimerization of crRNA and prime editing tracrRNA (petracrRNA) for Cas9 activity is guided by RNA annealing sequences (shown in purple) that are complementary. (c) Different designs of complementary sequences for the crRNA-petracrRNA interaction. (d–e) Editing efficiencies for prime editing (CTT insertion to HEK3 native genomic locus) using matching (d) or mixed (e) crRNA/petracrRNA pairs. CTT insertion efficiency using enhanced pegRNA (epegRNA) was used as the positive control (‘eCTT control’). The center and error bars are mean and standard deviations, respectively, from n = 3 transfection replicates.
-
Figure 1—source data 1
Source data (editing efficiencies) related to Figure 1 and figure supplements.
- https://cdn.elifesciences.org/articles/98110/elife-98110-fig1-data1-v1.xlsx
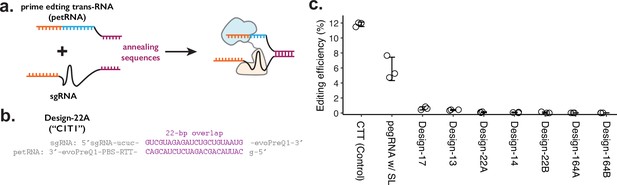
Testing the sgRNA:petRNA splitting strategy.
(a) Schematic of splitting functional prime editing guide RNA (pegRNA) into single-guide RNA (sgRNA) with extended sequence on 3’-end and petRNA with extended sequence on 5’-end for controlling dimerization. Annealing of two dimerization sequences would result in an active pegRNA. (b) Example design of dimerization sequence. Each design is named after the length of the reverse-complementary sequence (22 bp shown here). In both sgRNA and petRNA, evoPreQ1 RNA pseudoknot domains are added to reduce the degradation of the non-Cas9-bound RNA portion. (c) The editing efficiency was measured for seven different sgRNA:petRNA pairs along with prime editing control (pegRNA targeting HEK3 locus for CTT insertion at +0 position from the nick) and a single pegRNA construct with 22 bp RNA stem-loop between the sgRNA and reverse-transcription template (RTT) portion (pegRNA w/ SL) to test whether having extra RNA duplex affects the prime editing efficiency. The center and error bars are mean and standard deviations, respectively, from n = 3 transfection replicates.
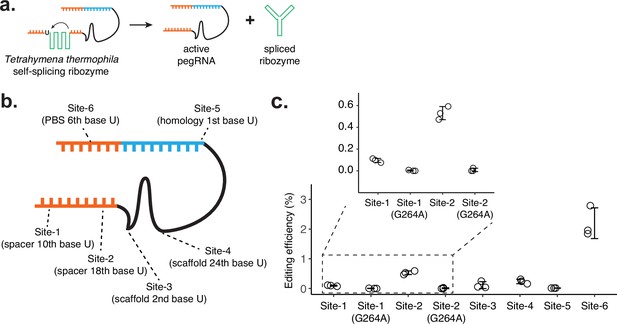
Testing the self-splicing ribozyme strategy.
(a) Schematic of self-splicing ribozyme strategy. Insertion of the ribozyme into functional prime editing guide RNA (pegRNA) would lead to self-splicing after RNA transcription, which would result in an active pegRNA and spliced ribozyme sequence. If this worked, the ribozyme could be further split into two functional parts, where extra dimerization domains can be added to control ribozyme function. (b) Six positions within the pegRNA (targeting HEK3 locus for CTT insertion) were tested for inserting the self-splicing ribozyme sequence. Ribozyme needs to start with a uridine base for splicing, which remains after the ribozyme is spliced out. (c) Editing efficiencies were measured for six ribozyme insertion sites within pegRNA. For Site-1 and Site-2, two additional designs were constructed with inactive ribozyme (G264A mutation). The center and error bars are mean and standard deviations, respectively, from n = 3 transfection replicates.

Controlling genome editing with constitutive or chemically induced protein-protein interactions.
(a) Schematic of P3 editing, which couples protein-protein interactions to genome editing. The binding of two proteins (e.g. MCP-tagged GCN4 epitope and LambdaN-tagged scFv) brings two RNA species (e.g. crRNA-MS2 and BoxB-petracrRNA) into close proximity to form an active prime editing guide RNA (pegRNA) complex, which binds to the prime editor for genome editing. (b) Normalized editing efficiencies measured with different protein pair interactions. Editing efficiencies were normalized to a consistent eCTT control included in each experiment (left-most). Transfection of filler GFP-expressing plasmid instead of proteins tagged with MCP/LambdaN was used to quantify the background levels of editing as a negative control (right-most). (c) Using chemically inducible dimerization to control genome editing. For example, FKBP and FRB domains dimerize in the presence of the small molecule rapamycin. (d) Normalized editing efficiencies measured for chemically induced genome editing. The center and error bars are mean and standard deviations, respectively, from n = 3 transfection replicates for both panels c and d.
-
Figure 2—source data 1
Source data (editing efficiencies) related to Figure 2 and figure supplements.
- https://cdn.elifesciences.org/articles/98110/elife-98110-fig2-data1-v1.xlsx
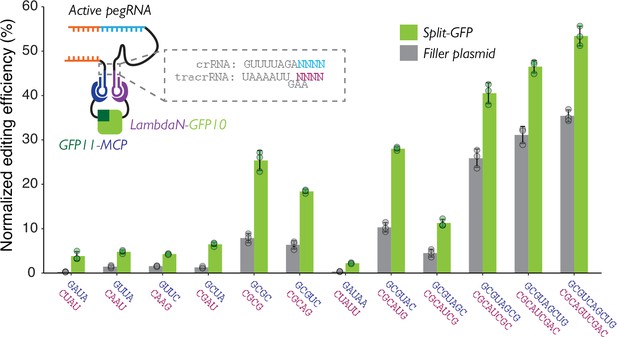
Characterizing linkers for crRNA-MS2 and BoxB-petracrRNA.
The upper four base-pairs of the Cas9-binding region of repeat:anti-repeat duplex were altered to generate 12 pairs of crRNA-MS2/BoxB-petracrRNA designs. The editing efficiency is normalized with the eCTT positive control included in each experiment (targeting HEK3 locus with CTT insertion at position +0 using standard enhanced prime editing guide RNA [epegRNA]), to control for variable transfection efficiencies. Two normalized editing efficiencies were measured for each pair of RNAs: one with tagged split-GFP to promote dual-RNA-guide formation, and one with standard, untagged GFP to measure the background editing level non-specific to protein-protein proximity. The center and error bars are mean and standard deviations, respectively, from n = 3 transfection replicates.
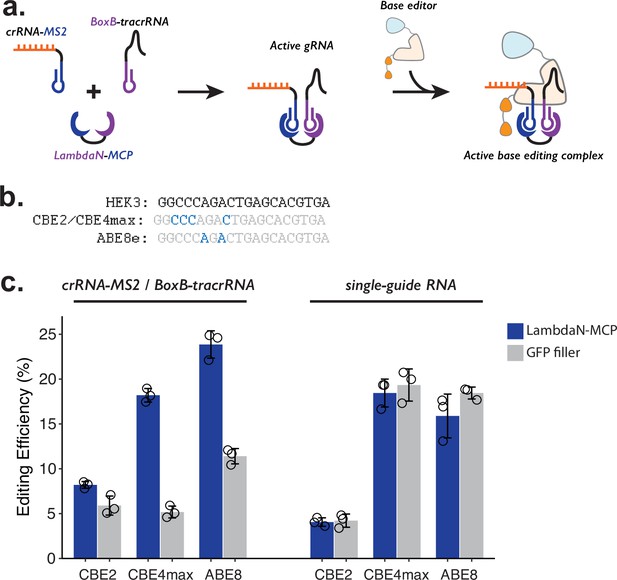
Testing P3 editing strategy in the context of base editing.
(a) Schematics of P3 editing strategy adapted to base editing. The LambdaN-MCP fusion protein will bring crRNA-MS2 and BoxB-tracrRNA in close proximity to aid gRNA complex formation, which then binds to the base editor for genome editing. (b) Cytosine and adenine residues targeted by base editing are highlighted within the target HEK3 sequence. (c) Editing efficiency measured with P3 editing (left) and standard sgRNA (right). For each base editor and guide-RNA combination, editing efficiencies were measured with either LambdaN-MCP expressing plasmid (blue) or a filler plasmid expressing GFP (gray). The center and error bars are mean and standard deviations, respectively, from n = 3 transfection replicates.
-
Figure 3—source data 1
Source data (editing efficiencies) related to Figure 3 and figure supplements.
- https://cdn.elifesciences.org/articles/98110/elife-98110-fig3-data1-v1.xlsx
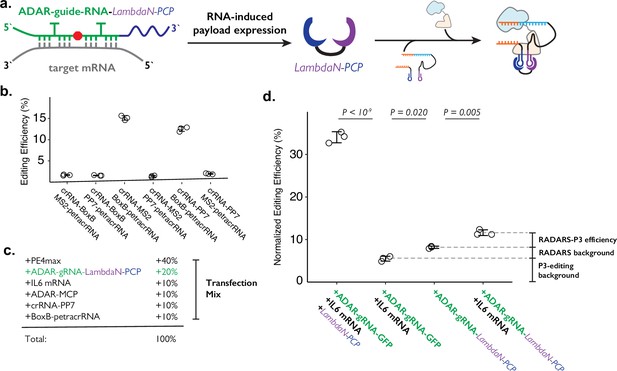
Combining ADAR-based RNA sensors with P3 editing.
(a) Schematics of combining ADAR-based RNA sensors with P3 editing. ADAR-guide-RNA senses target RNA by forming extended RNA duplex based on reverse-complementary sequence, which is a substrate for RNA editing by ADAR. RNA editing converts the in-frame stop codon into a sense codon, which then starts the expression of cargo protein. In combination with P3 editing, the cargo protein will be LambdaN-PCP, which will then help prime editing complex formation by bringing crRNA-PP7 and BoxB-petracrRNA together. (b) The six pairs of crRNA/petracrRNA with three different RNA aptamers (BoxB, MS2, and PP7) were tested for P3 editing. Pairing of crRNA-PP7 and BoxB-petracrRNA is similarly compatible with P3 editing as pairing of crRNA-MS2 and BoxB-petracrRNA, and is preferable in this context as it avoids using the MS2 RNA element which is used in RADARS strategy for the ADAR-based RNA sensing. The center and error bars are mean and standard deviations, respectively, from n = 3 transfection replicates. (c) Composition of transfection condition with six expression plasmid. The percentage is based on the mass amount of plasmid added to the transfection mix. (d) The normalized editing efficiency was measured to characterize the efficiency and specificity of ADAR-based RNA sensing and P3 editing strategy. p-Values were obtained using a two-tailed Student’s t-test with Bonferroni correction. The center and error bars are mean and standard deviations, respectively, from n=3 transfection replicates.
-
Figure 4—source data 1
Source data (editing efficiencies) related to Figure 4 and figure supplements.
- https://cdn.elifesciences.org/articles/98110/elife-98110-fig4-data1-v1.xlsx
Tables
| Reagent type (species) or resource | Designation | Source or reference | Identifiers | Additional information |
|---|---|---|---|---|
| Strain, strain background (E. coli) | NEB C3040H Competent cells | NEB | C3040H | |
| Cell line (Homo-sapiens) | HEK293T | ATCC | CRL-3216 | |
| Transfected construct (synthetic) | PE4max | Addgene | Addgene_174828 | |
| Transfected construct (synthetic) | pU6-pegRNA-GG-acceptor | Addgene | Addgene_132777 | |
| Transfected construct (synthetic) | pU6-crRNA-MS2 | Addgene | Addgene_207624 | |
| Transfected construct (synthetic) | pU6-BoxB-petracrRNA | Addgene | Addgene_207625 | |
| Transfected construct (synthetic) | pCMV-LambdaN-MCP | Addgene | Addgene_207626 | |
| Transfected construct (synthetic) | pCMV-LambdaN-NbALFA | Addgene | Addgene_207627 | |
| Transfected construct (synthetic) | pCMV-ALFA-MCP | Addgene | Addgene_207628 | |
| Commercial assay or kit (cloning) | T4 DNA ligase | NEB | M0202S | |
| Commercial assay or kit (cloning) | BsaI-HF-v2 | NEB | R3733S | |
| Commercial assay or kit (cloning) | NEBuilder HiFi DNA assembly master mix | NEB | E2621S | |
| Commercial assay or kit (plasmid purification) | Qiagen miniprep | Qiagen | 27106 | |
| Commercial assay or kit (transfection reagent) | Lipofectamine 3000 | Thermo Fisher | L3000001 | |
| Commercial assay or kit (PCR) | KAPA2G Robust 2 x Hotstart mix | Roche | KK5702 |
Additional files
-
Supplementary file 1
Nucleic acid sequences used in this study.
- https://cdn.elifesciences.org/articles/98110/elife-98110-supp1-v1.xlsx
-
MDAR checklist
- https://cdn.elifesciences.org/articles/98110/elife-98110-mdarchecklist1-v1.docx
-
Source data 1
Combined table including source data (editing efficiencies) related to all figures and figure supplements in this article.
- https://cdn.elifesciences.org/articles/98110/elife-98110-data1-v1.xlsx





