Multi-tissue network analysis reveals the effect of JNK inhibition on dietary sucrose-induced metabolic dysfunction in rats
Figures
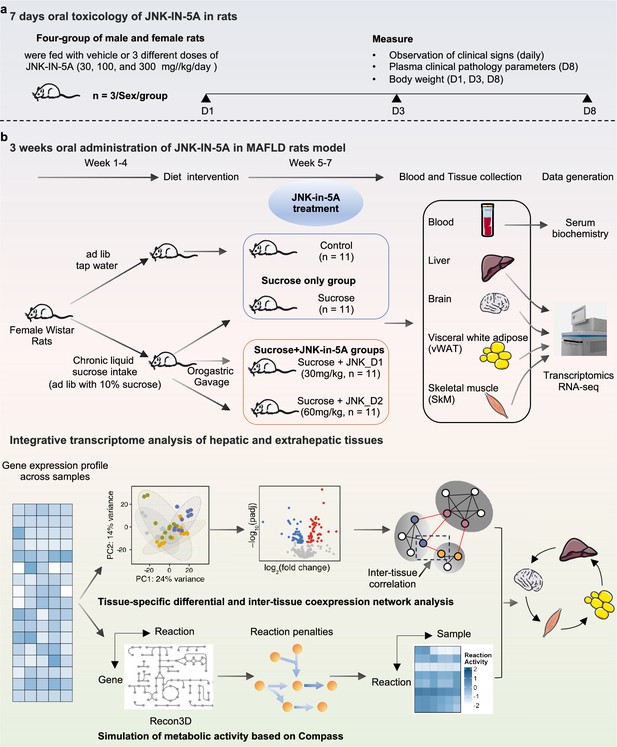
Schematic representation of experimental setup.
Study groups included the healthy control group received tap water, the sucrose group received 10% sucrose water, the sucrose+JNK_D1 group received 10% sucrose and 30 mg/kg/d JNK-IN-5A, and the sucrose+JNK_D2 group received 10% sucrose and 60 mg/kg/d JNK-IN-5A (N=44, n=11 per group).
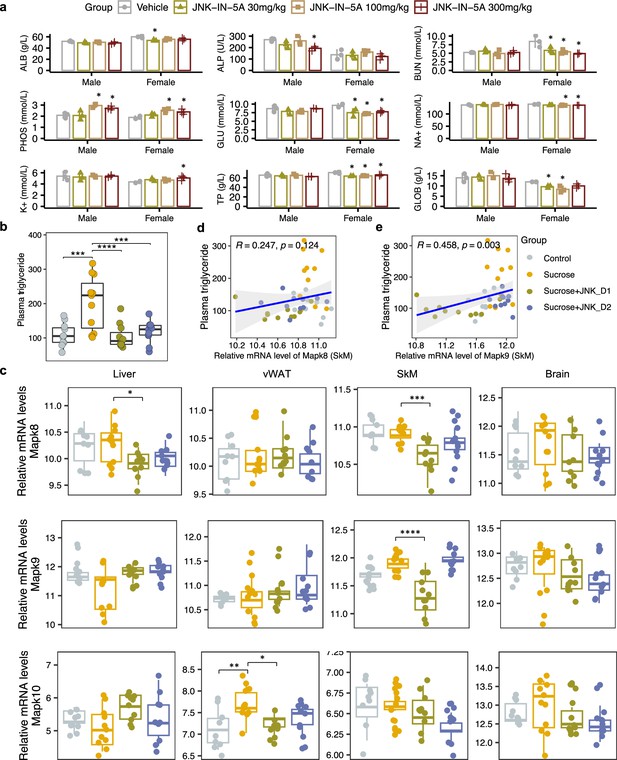
Sucrose consumption and JNK-IN-5A treatment exhibit distinct effects on the mRNA expression of genes encoding c-Jun N-terminal kinase (JNK) isoforms in the major metabolic tissues.
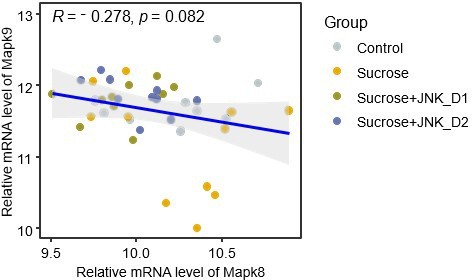
(a) The acute effect of JNK-IN-5A in rats (n=3/Sex/Group). Statistically significant changed clinical chemistry parameters were presented as mean ± standard deviation (SD), see also Supplementary file 1. ALB: albumin; ALP: alkaline phosphatase; BUN: blood urea nitrogen; PHOS: phosphate; GLU: glucose; K+: potassium; Na+: sodium; TP: total Protein; GLOB: globulin. Statistical significance was assessed by one-way ANOVA testing followed by Tukey’s multiple comparisons post-test or Shapiro–Wilk testing followed by Dunn’s post-hoc test, as appropriate, <i>p-value <0.05 is considered as statistical significance. (b) Boxplot showing the levels of plasma triglycerides in control, sucrose, sucrose+JNK_D1, and sucrose +JNK_D2 rats. Statistical significance was assessed by one-way ANOVA testing followed by Tukey’s multiple comparisons post-test or the Kruskal-Wallis test followed by Dunn’s multiple comparisons post-test, as appropriate, <i>p-value <0.05 is considered as statistical significance. *p<0.05, **p<0.01, ***p<0.001, ****p<0.0001. (c) Boxplot showing the relative mRNA expression of Mapk8, Mapk9, and Mapk10 in the studied tissues. The count-based abundance of genes was transformed using the vst function and the Benjamini-Hochberg adjusted p-value (p.adj) is derived from DESeq2 (Love et al., 2014). *p.adj <0.05, **p.adj <0.01, ***p.adj <0.001, ****p.adj <0.0001. (d) Correlation between the relative mRNA expression of Mapk8 and (e) Mapk9 in skeletal muscle tissue (SkM) and plasma triglycerides level.
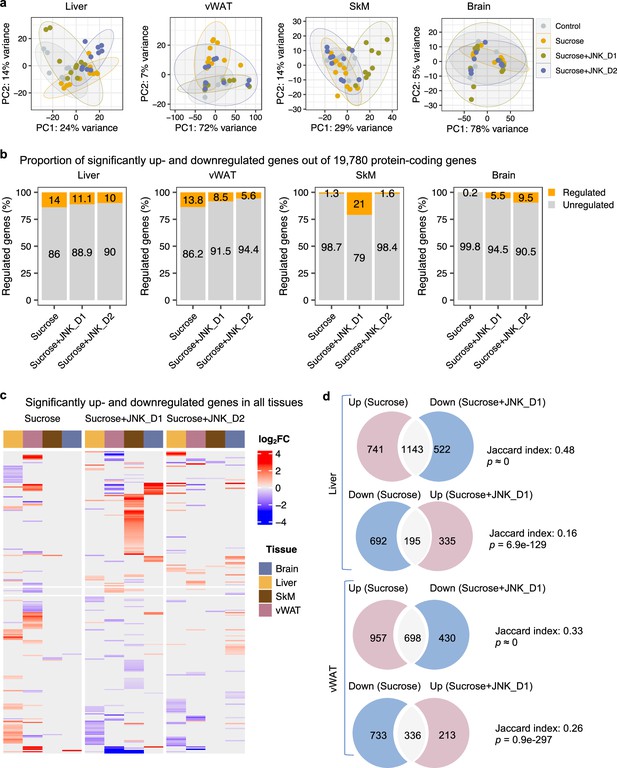
System-wide transcriptomics profiling revealed tissue-specific metabolic rewiring by sucrose consumption and JNK-IN-5A treatment.
(a) Principal component analysis (PCA) of liver, visceral white adipose tissue (vWAT), skeletal muscle (SkM), and brain transcriptome data showing global gene expression profiles in control (n=9), sucrose (n=11), sucrose+JNK_D1 and sucrose+JNK_D2 (n=10/per group). Each data point represents a sample in the respective colored group. (b) Bar graphs represent the percentage of differential expressed genes (or regulated genes) associated with sucrose (sucrose vs. control), sucrose+JNK_D1 (sucrose+JNK_D1 vs. sucrose), or sucrose+JNK_D2 (sucrose+JNK_D2 vs. sucrose) in all protein-coding genes (N=19,780) for each tissue. (c) Heatmap of all genes differentially upregulated (red) and downregulated (blue) in three pairwise comparisons for sucrose (sucrose vs. control), sucrose+JNK_D1 (sucrose+JNK_D1 vs. sucrose), and sucrose+JNK_D2 (sucrose+JNK_D2 vs. sucrose) (adjusted p-value, p.adj<0.01) for each tissue. Column annotation represents tissues. (d) Venn diagram showing the overlap between significantly regulated genes response to sucrose consumption and JNK_D1 treatment.
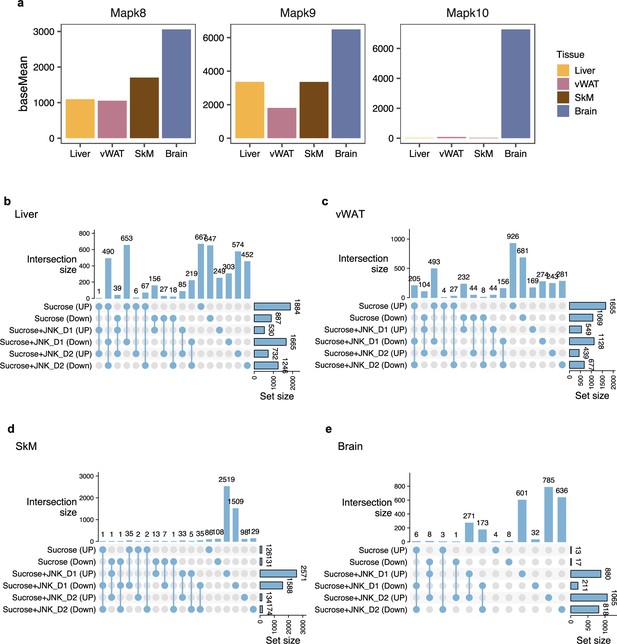
Intersection between differentially expressed genes in different tissues.
(a) Gene expression pattern of c-Jun N-terminal kinase (JNK) isoforms (b–e) UpSet plot showing the number of shared differentially expressed genes (DEGs) (adjusted-p<0.01) associated with sucrose consumption and JNK-IN-5A treatment in the liver, visceral white adipose tissue (vWAT), skeletal muscle tissue (SkM), and brain tissues in the experimental rats. The histogram bar graph (right) shows the total number of DEGs in each comparison corresponding. The histogram bar graph (top) shows the number of DEGs shared among pairwise comparisons. Connected lines indicate what comparisons are sharing DEGs.
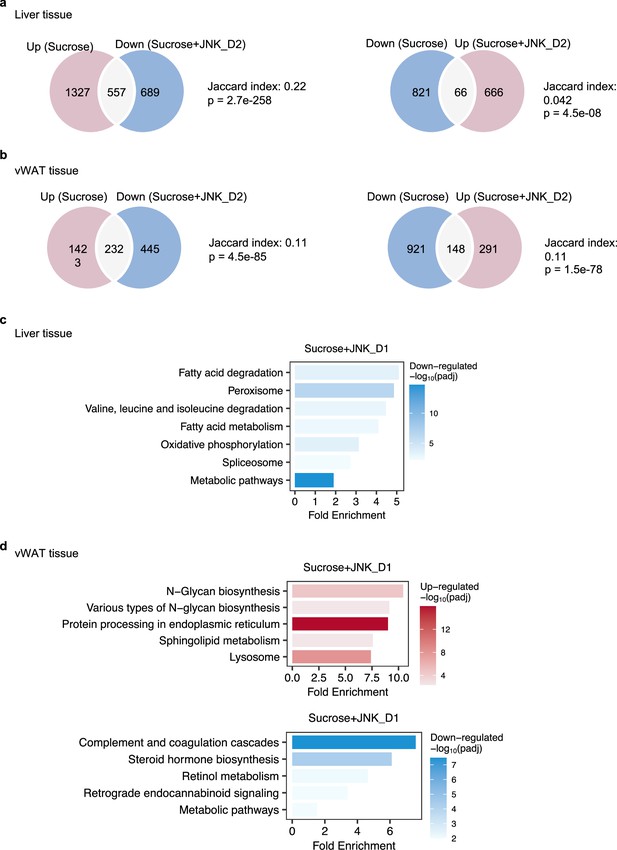
Transcriptional changes in the liver and adipose tissues in response to JNK-IN-5A treatment.
(a) Venn diagram showing the overlap between significantly regulated genes response to sucrose consumption and JNK_D2 treatment in liver and (b) in visceral white adipose tissue (vWAT). (c) Top 10 enriched KEGG pathways enriched by reversed genes after JNK_D1 treatment in liver and (d) in vWAT.
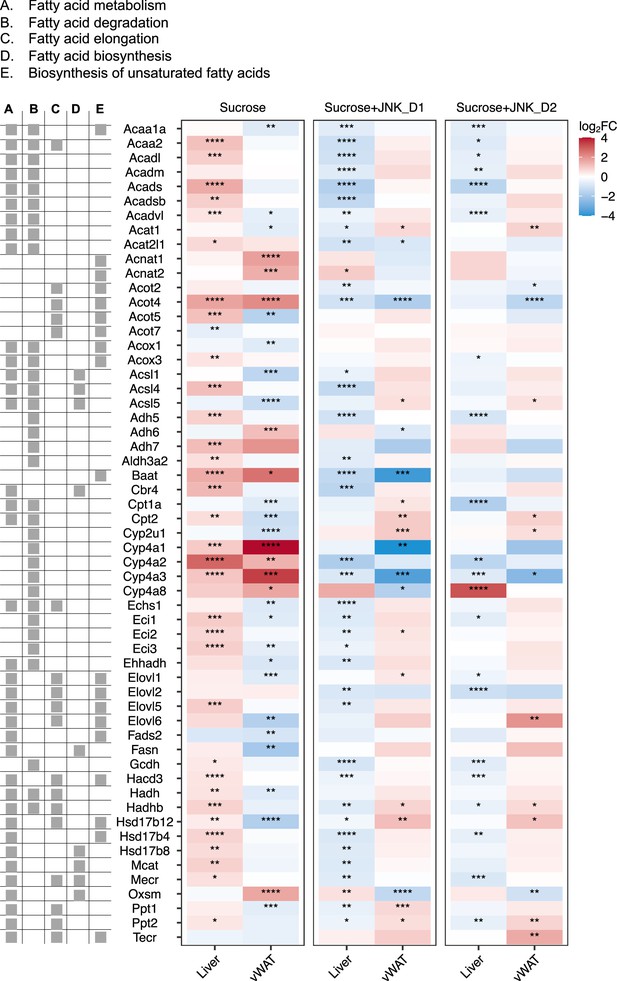
Relative changes of genes involved in fatty acid degradation, elongation, biosynthesis, and metabolism in response to sucrose consumption and JNK-IN-5A treatment in the tissues.
The BH-adjusted p-value (p.adj) is derived from DESeq2 (Love et al., 2014). *p.adj<0.05, **p.adj<0.01, ***p.adj<0.001, ****p.adj<0.0001.
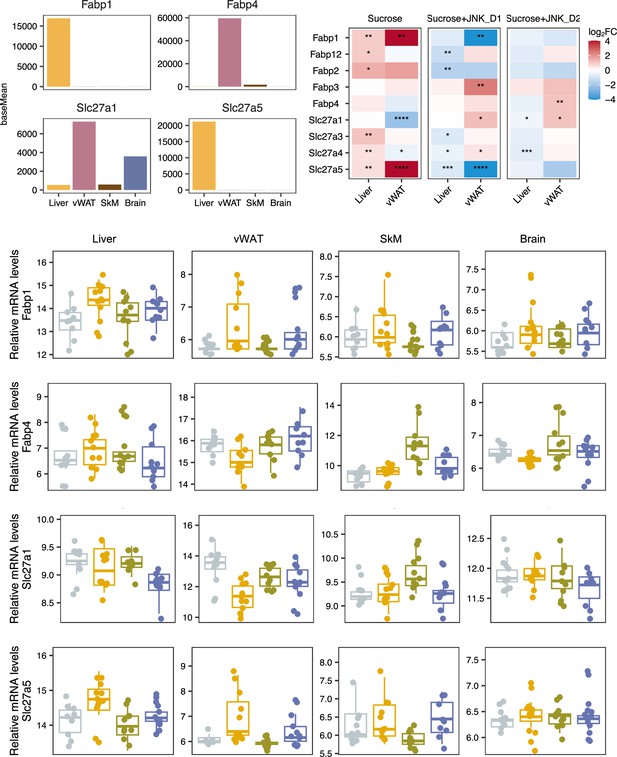
Relative changes of genes involved in fatty acid uptake in response to sucrose consumption and JNK-in-5A treatment in the tissues.
The BH-adjusted p-value (p.adj) is derived from DESeq2 (Love et al., 2014). *p.adj<0.05, **p.adj<0.01, ***p.adj<0.001, ****p.adj<0.0001.
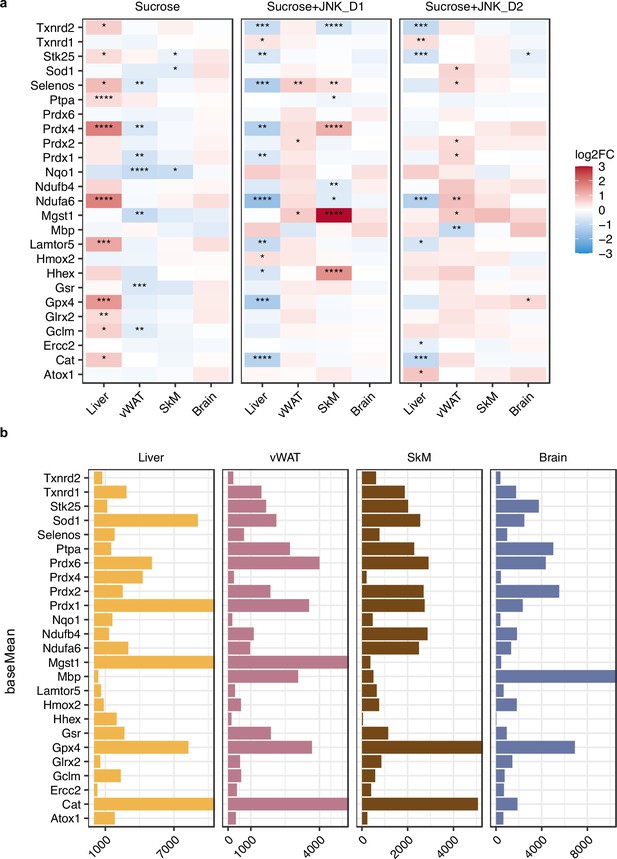
Relative changes of genes involved in redox homeostasis in response to sucrose consumption and JNK-in-5A treatment in the tissues.
The BH-adjusted p-value (p.adj) is derived from DESeq2 (Love et al., 2014). *p.adj<0.05, **p.adj<0.01, ***p.adj<0.001, ****p.adj<0.0001.
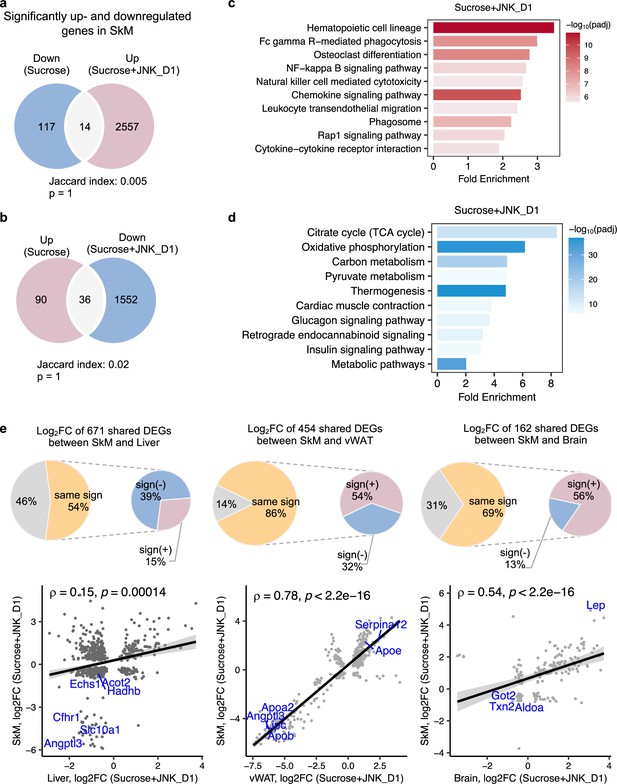
c-Jun N-terminal kinase (JNK) inhibition in skeletal muscle (SkM) correlated with the regulation of energy metabolism in the liver and adipose tissues.
(a, b) Venn diagram showing the overlap between significantly regulated genes response to sucrose consumption and INK_D1 treatment in SkM. (c, d) Functional annotation of up- (red) and down-regulated (blue) genes in JNK_D1-treated SkM with a Benjamini Hochberg adjusted p-value <0.05. (e) Correlation of log2(Fold change) for shared differentially expressed genes (DEGs) between the SkM and Liver, visceral white adipose tissue (vWAT), or Brain after JNK_D1 treatment.
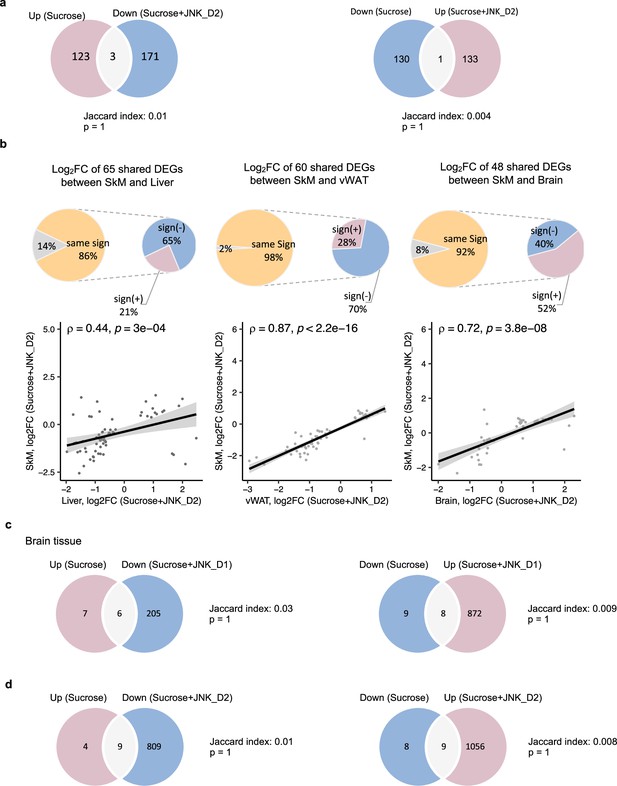
Transcriptional changes in response to JNK-IN-5A treatment.
(a) Venn diagram showing the overlap between significantly regulated genes response to sucrose consumption and JNK_D2 treatment in skeletal muscle (SkM). (b) Correlation of log2(Fold change) for shared differentially expressed genes (DEGs) between the SkM and Liver, visceral white adipose tissue (vWAT), or Brain after JNK_D2 treatment. (c) Venn diagram showing the overlap between significantly regulated genes response to sucrose consumption and JNK_D1 treatment as well as (d) JNK_D2 treatment in the Brain, respectively.
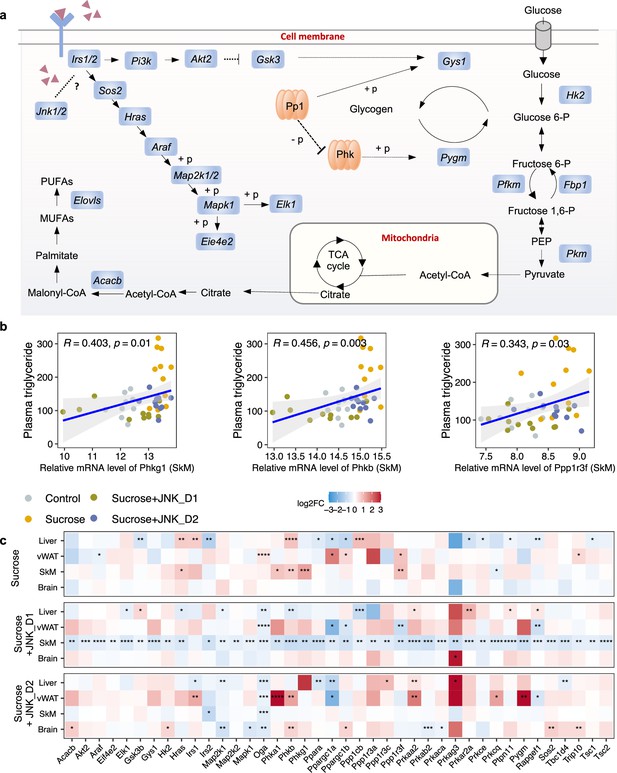
c-Jun N-terminal kinase (JNK) inhibition regulates insulin signaling-related genes in skeletal muscle (SkM).
(a) A representative diagram of metabolic pathways associated with insulin resistance and insulin signaling was found to be largely inhibited by JNK_D1 treatment. (b) Correlation between the relative mRNA expression of Phkg1, Phkb, and Ppp1r3f (those three genes are also significantly upregulated in sucrose-feeding only group) in SkM and plasma triglycerides level. (c) Relative changes of genes involved in insulin resistance and insulin signaling pathways in response to sucrose consumption and JNK-IN-5A treatment in the tissues. The BH-adjusted p-value (p.adj) is derived from DESeq2 (Love et al., 2014). *p.adj<0.05, **p.adj<0.01, ***p.adj<0.001, ****p.adj<0.0001.
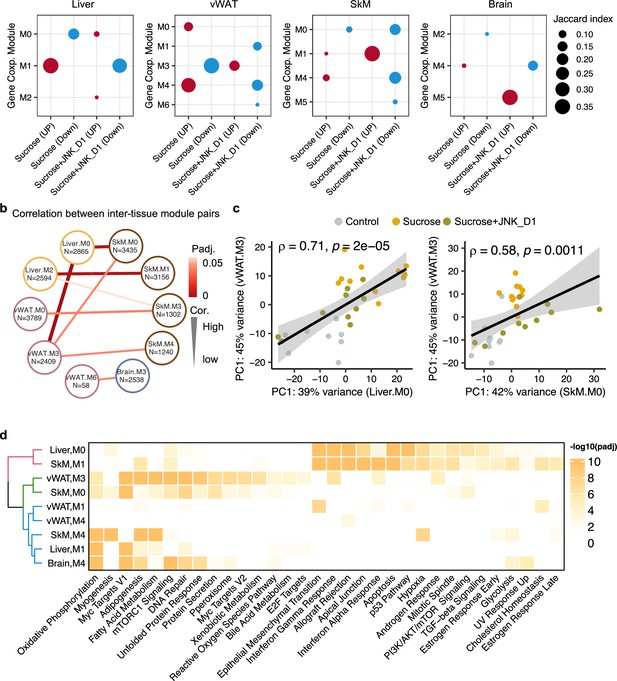
Inter-tissue network analysis identifies JNK-IN-5A inhibition-associated molecular mechanism.
(a) Significantly enriched modules (hypergeometric test, <0.05) by the differentially expressed genes related to sucrose, sucrose+JNK_D1, and sucrose+JNK_D1 in each tissue. (b) Dot-plot showing the significant correlation among inter-tissue module pairs. The size and color of the connected line are proportional to the correlation coefficient and statistical significance indicated by the adjusted p-value. The module pairs with Benjamini Hochberg adjusted p-value (p.adj)<0.05 are presented. (c) Spearman coefficient correlation of the first component from principal component analysis (PCA) analyses on gene expression of module pairs. (d) Significantly enriched MSigDB hallmark gene sets by each module.
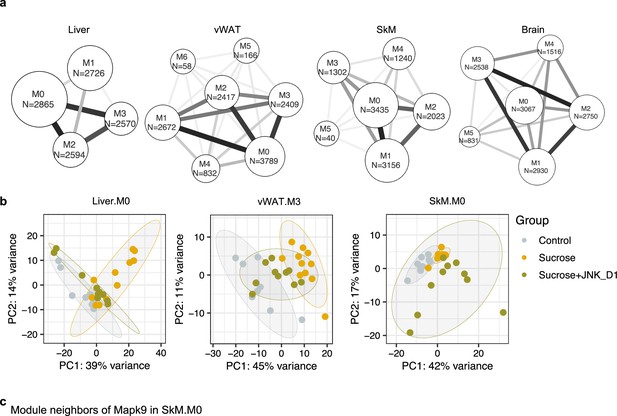
Tissue-specific gene co-expression network analyses.
(a) The number of gene modules are identified from the tissue-specific co-expression networks. The text on the module indicates the module name and the number of genes of individual modules in each tissue. The edges between the clusters were aggregations of the inter-cluster edges. (b) Principal component analysis (PCA) of Liver.M0, vWAT.M3, and SkM.M0 gene expression data. (c) Module neighbors of Mapk9 in SkM.M0.
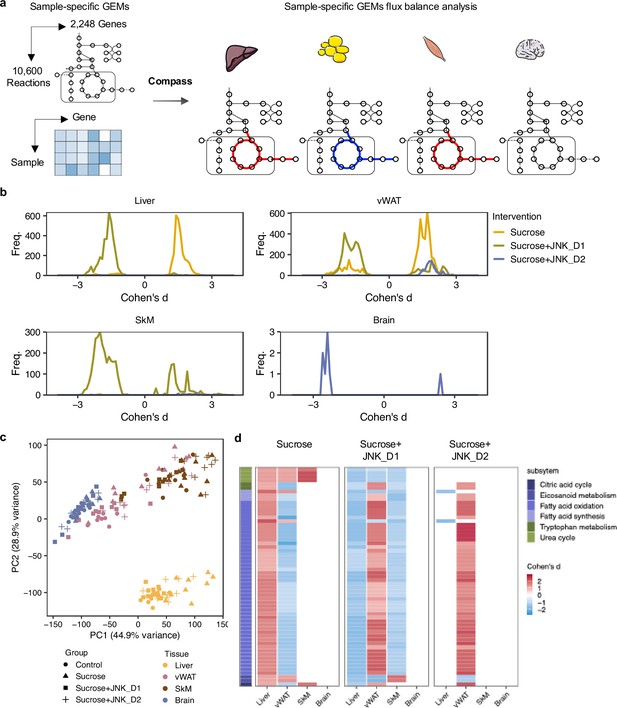
Metabolic modeling highlights metabolic reprogramming linked to JNK-IN-5A inhibition.
(a) Simulation of metabolic activity based on Compass (Wagner et al., 2021) and Recon3D (Brunk et al., 2018). (b) Distribution of Cohen’s d statistic for differential metabolic reactions in response to sucrose, sucrose+JNK_D1 and sucrose+JNK_D2 in each tissue, see also Supplementary file 8. (c) Principal component analysis of the Compass score of metabolic reaction potential activity scores showing the metabolic heterogeneity in response to sucrose ingestion and JNK-IN-5A treatment at the tissue levels, for principal components 1 and 2. (d) Heatmap showing the tissue-level differential activity of metabolic reactions in response to sucrose intake and JNK-IN-5A treatment. Reactions are partitioned by Recon3 pathways (Brunk et al., 2018) and colored by the sign of their Cohen’s d statistic derived from different contrasts: sucrose vs control, sucrose+JNK_D1 vs sucrose, or sucrose+JNK_D2 vs sucrose, respectively. Abbreviations: vWAT, visceral white adipose tissue; SkM, skeletal muscle.
Tables
Metabolites associated with sucrose overconsumption in MASLD.
| Study | Intervention(s) and duration | Metabolites | Metabolism |
|---|---|---|---|
| Fujii et al., 2024 [1]. | Male rats fed a 10% sucrose solution for 16 days | Decreased blood acetate and butyrate levels | Gut microbial metabolism, Lipid metabolism |
| Sun et al., 2021 [2]. | Male Wistar rats fed high-sucrose diet for 4 weeks | Decreased butyrate and formate in the cecal content | |
| Song et al., 2024 [3]. | Male Wistar rats fed a high sucrose diet for 4 weeks | Decreased acetate and butyrate; increased succinate in cecal content | |
| Ramos-Romero et al., 2019 [4]. | Male Wistar rats were fed a 35% sucrose solution for 24 weeks. | Increased uric acid in urine | Urea cycle |
Field, 2023 [5]. | Male C57BL/6J mice were fed either low-fat diet or high-fat diet with 30% sucrose water for 8 weeks | Elevated serum and kidney erythritol concentration | Carbohydrate metabolism |
| Beckmann et al. 2015 [6]. | Healthy females (n=90) consumed 0,50 , or 100 g sucrose in water, followed by urine and blood sampling at 0,3 , and 24 hours. | Increased erythronic acid in plasma | Carbohydrate metabolism |
| He et al., 2023 [7]. | Male C57BL/6J mice were fed a high sucrose diet (30% sucrose in drinking water) for 24 weeks | Decreased muricholic acid level in the liver, cecal and colon content. increased hyocholic acid levels in the serum. | Bile acid metabolism |
| Stephenson et al. 2022 [8]. | Mice fed 10% sucrose solution in drinking water for 12 weeks | Increased triglyceridebound oleate, palmitate, and stearate in liver; Mixed alteration in serum bile acids pool (sex and treatment interaction effect) | Lipid metabolism and bile acid metabolism |
| Mock et al. 2017 [9]. | Female rats fed 13% sucrose solutions for 8 weeks | Increased palmitoleic acid in gonadal and retroperitoneal fat pads; higher serum triglyceride | Lipid metabolism |
| Oztūrk et al. 2022 [10]. | Wistar male rats fed 10% sucrose in drinking for 3 months | Decreased serum levels of kynurenic acid and kynurenine | Tryptophan metabolism |
| Gariani, Karim, et al. 2016 [11]. | Male C57BL/6J mice were fed with a Western high-fat and highsucrose | Decreased NAD* levels in the liver | Fatty acid oxidation |
| Togo et al., 2019 [12]. | C57BL/6J mice fed liquid (50% by weigh) or solid sucrose for 8 weeks | Elevated hepatic fat | Lipid metabolism |
Additional files
-
Supplementary file 1
Summary of body weight and clinical pathological parameters for 7 d of oral toxicology of JNK-IN-5A in rats.
- https://cdn.elifesciences.org/articles/98427/elife-98427-supp1-v2.xlsx
-
Supplementary file 2
Summary of body weight and clinical pathological parameters for 3 wk oral administration of JNK-IN-5A in metabolic dysfunction‐associated fatty liver disease (MAFLD) rats model.
- https://cdn.elifesciences.org/articles/98427/elife-98427-supp2-v2.xlsx
-
Supplementary file 3
Passive avoidance data.
- https://cdn.elifesciences.org/articles/98427/elife-98427-supp3-v2.xlsx
-
Supplementary file 4
Locomotor activity data.
- https://cdn.elifesciences.org/articles/98427/elife-98427-supp4-v2.xlsx
-
Supplementary file 5
Elevated plus maze data.
- https://cdn.elifesciences.org/articles/98427/elife-98427-supp5-v2.xlsx
-
Supplementary file 6
Result of differential expression analysis for all relevant comparisons of liver, white adipose tissue, skeletal muscle, and brain in the study, related to Figures 2—4.
- https://cdn.elifesciences.org/articles/98427/elife-98427-supp6-v2.xlsx
-
Supplementary file 7
Gene membership in certain modules identified from the tissue-specific co-expression networks, which includes samples from control, sucrose, and sucrose+JNK_D1 groups, related to Figure 6.
- https://cdn.elifesciences.org/articles/98427/elife-98427-supp7-v2.xlsx
-
Supplementary file 8
Results of genome-scale metabolic modeling based on Compass and Recon3D, related to Figure 7.
- https://cdn.elifesciences.org/articles/98427/elife-98427-supp8-v2.xlsx
-
Supplementary file 9
Metabolites associated with sucrose overconsumption in MASLD.
- https://cdn.elifesciences.org/articles/98427/elife-98427-supp9-v2.docx
-
MDAR checklist
- https://cdn.elifesciences.org/articles/98427/elife-98427-mdarchecklist1-v2.docx