Phosphodiesterase 1A physically interacts with YTHDF2 and reinforces the progression of non-small cell lung cancer
Figures
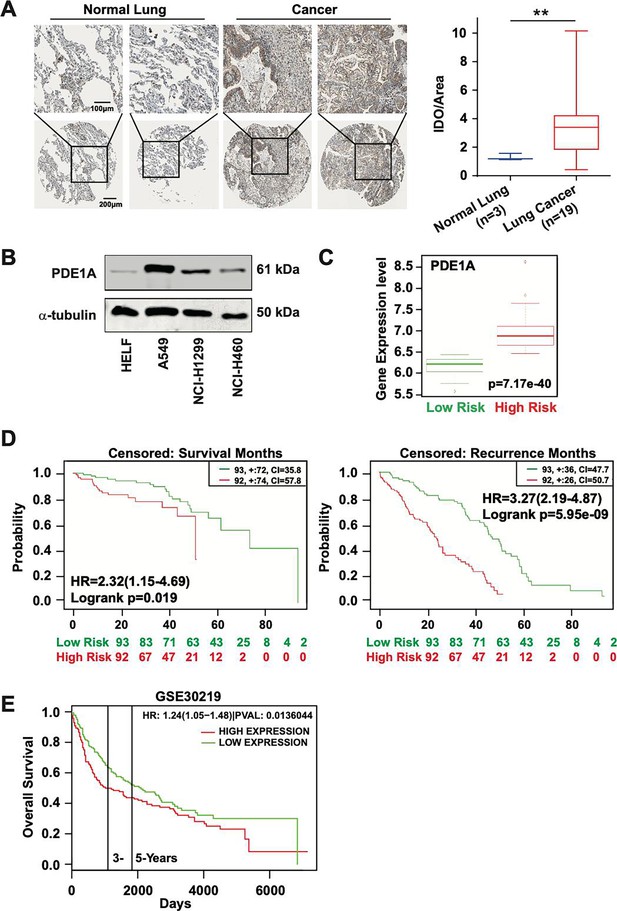
High expression of phosphodiesterase 1A (PDE1A) predicts a poor prognosis of lung cancer patients.
(A) The expression of PDE1A was detected in non-small cell lung cancer (NSCLC) and normal lung tissue. It was obtained from The Human Protein Atlas (https://www.proteinatlas.org/). IOD/area means integral optical density/area. t-Test was used to compare the difference between lung cancer and normal lung groups, **P < 0.01. (B) The expression of PDE1A in human lung fibroblasts (HELF), A549, NCI-H1299, and NCI-H460 cells was detected by western blot. (C and D) Box plot analysis of the PDE1A messenger RNA (mRNA) levels in clinical lung cancer tissue samples. It was collected, and statistical analyses were performed from SurvExpress (http://bioinformatica.mty.itesm.mx:8080/Biomatec/SurvivaX.jsp). Gene: PDE1A; Access database numbers: Chitale lung (n=185); Censored: recurrence months or survival months. (E) It was collected from PROGgene V2 Prognostic Database (http://www.progtools.net/gene/index.php). Survival analysis is done using backend R script which employs R library ‘survival’ to perform Cox proportional hazards analysis (function ‘coxph’) and to plot prognostic plots (function ‘survfit’). Single-user input genes: PDE1A; Cancer type: LUNG; Survival measure: death; Bifurcate gene expression at: median; GSE30219-Off-context gene expression in lung cancer identifies a group of metastatic-prone tumors.
-
Figure 1—source data 1
Raw images for western blot shown in Figure 1B (labelled).
- https://cdn.elifesciences.org/articles/98903/elife-98903-fig1-data1-v1.zip
-
Figure 1—source data 2
Raw images for western blot shown in Figure 1B.
- https://cdn.elifesciences.org/articles/98903/elife-98903-fig1-data2-v1.zip
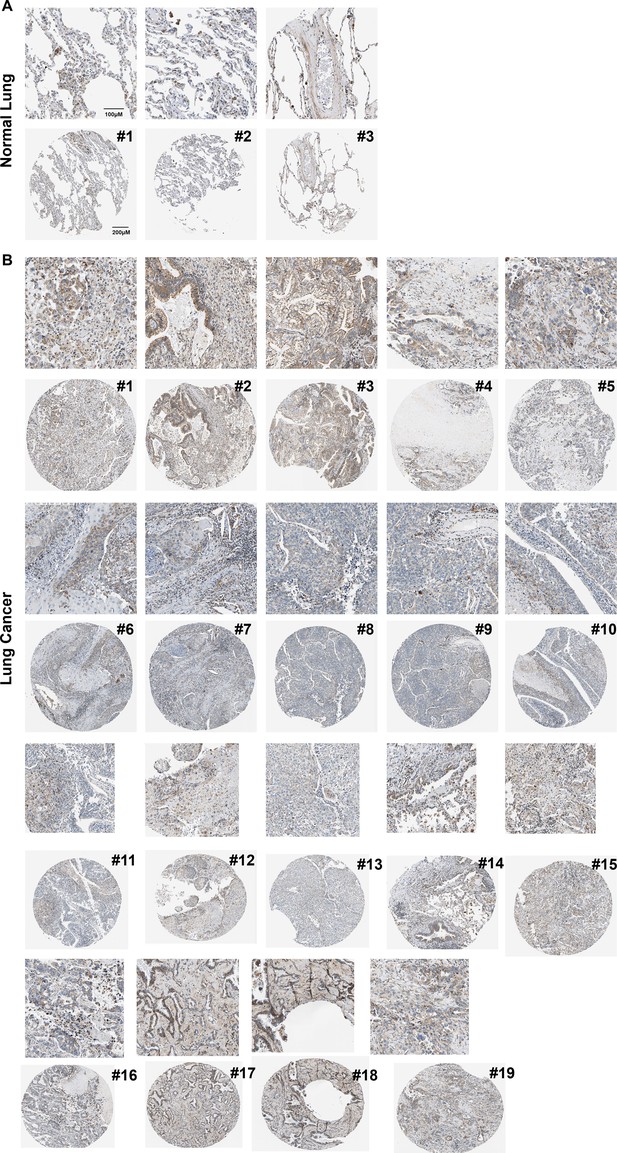
The expression of phosphodiesterase 1A (PDE1A) in normal lung (A) and lung cancer (B).
It was obtained from The Human Protein Atlas (https://www.proteinatlas.org/).
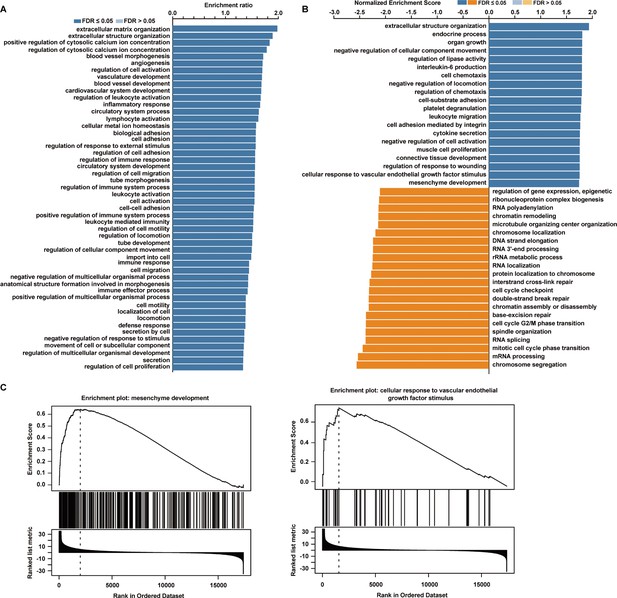
The biological processes related to phosphodiesterase 1A (PDE1A) in non-small cell lung cancer (NSCLC) are predicted using LinkedOmics.
Data collected from LinkedOmics (https://www.linkedomics.org/admin.php) are shown. Overrepresentation enrichment analysis (ORA) (A) and gene set enrichment analysis (GSEA) (B) were performed to analyze the biological processes related to PDE1A in NSCLC. Sample cohort: TCGA_NSCLC; Institute: UNC; Data type: RNAseq; Platform: HiSeq RNA; Attribute: PDE1A; Statistical methods: Spearman’s correlation test; Patients: 515; Tools: ORA and GSEA; Gene Ontology analysis: biological process. (C) GSEA demonstrated that PDE1A participated in mesenchyme development and the cellular response to VEGF stimulus.
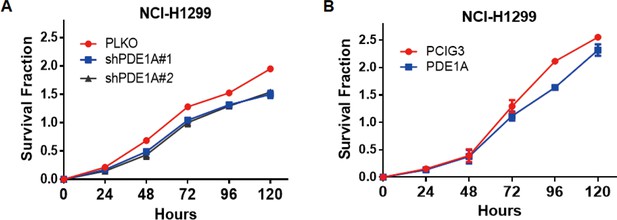
The expression of phosphodiesterase 1A (PDE1A) does not affect the proliferation of non-small cell lung cancer (NSCLC) cells.
(A–B) NSCLC cells were transfected with PDE1A siRNA or control siRNA (A) and empty vector or PDE1A plasmid (B) for 24 hr in six-well plates, then transferred to 96-well plates for the indicated times, and finally subjected to sulforhodamine B (SRB) assay (n=3).
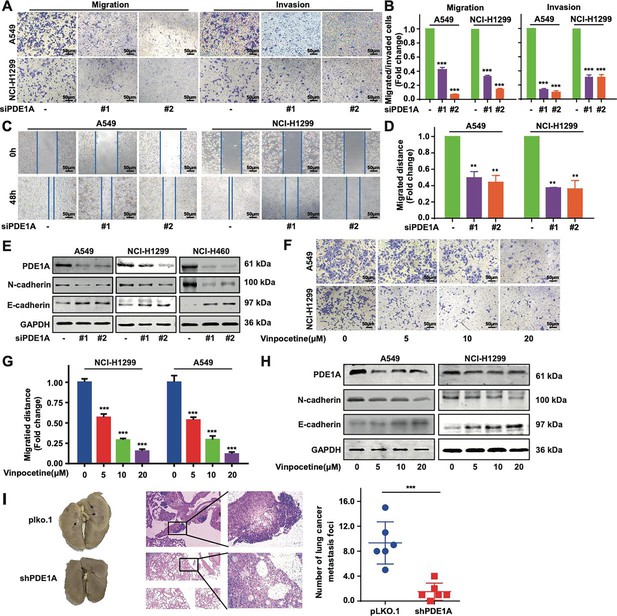
Phosphodiesterase 1A (PDE1A) knockdown suppresses the metastasis of non-small cell lung cancer (NSCLC) cells.
(A–B) NSCLC cells were transfected with control siRNA and PDE1A siRNA for 24 hr. Cells were transferred to Transwell chambers without or with a Matrigel coating on the insert membrane, and the cell migrative and invasive abilities were determined, respectively (n=3). (C–D) NSCLC cells were transfected with control siRNA and PDE1A siRNA for 24 hr, and the wound healing assay was established in NSCLC cells (n=3). (E) NSCLC cells were transfected with control siRNA and PDE1A siRNA for 48 hr, and the expression of indicated proteins was detected. (F–G) NSCLC cells were treated with DMSO or vinpocetine (5, 10, 20 µM) for 24 hr, and the migrative ability of treated NSCLC cells was determined using the Transwell assay for 24 hr (n=3). (H) NSCLC cells were treated with DMSO or vinpocetine (5, 10, 20 µM) for 24 hr, and the expression of indicated proteins was determined. (I) The pulmonary metastatic nodules were stained using H&E staining and counted in nude mice harboring NCI-H1299 cells transfected with PDE1A shRNA and control shRNA (n=6). **P < 0.01, ***P < 0.001.
-
Figure 2—source data 1
Raw images for western blots shown in Figure 2 (labelled).
- https://cdn.elifesciences.org/articles/98903/elife-98903-fig2-data1-v1.zip
-
Figure 2—source data 2
Raw images for western blots shown in Figure 2.
- https://cdn.elifesciences.org/articles/98903/elife-98903-fig2-data2-v1.zip
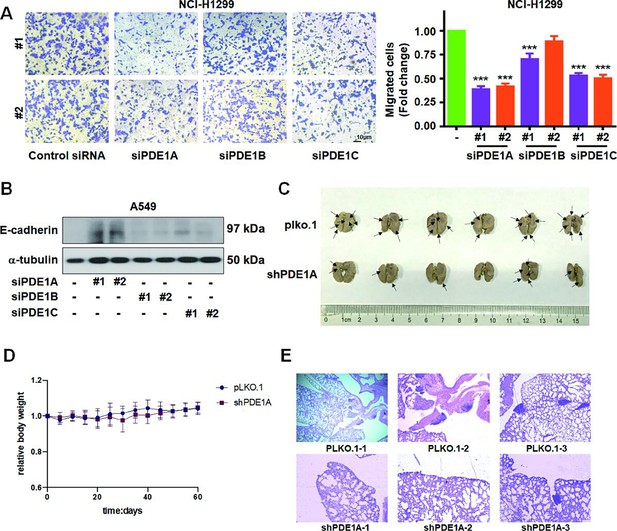
Phosphodiesterase 1A (PDE1A) silence suppresses the metastasis of non-small cell lung cancer (NSCLC) cells.
(A) NSCLC cells were transfected with control siRNA, siPDE1A, siPDE1B, and siPDE1C for 24 hr, and cells were transferred to Transwell chambers without a Matrigel coating on the insert membrane, and the cell migration was determined after 24 hr, (n=3). (B) NSCLC cells were transfected with indicated siRNA for 48 hr, and the expression of the indicated proteins was detected. (C) The lungs of nude mice were separated and stained with Bouin’s solution, and the pulmonary metastatic nodules were counted. (D) The relative body weight of each group in the in vivo metastasis studies in Figure 2I, (n=6). (E) The lungs in nude mice were stained using H&E. ***P < 0.001.
-
Figure 2—figure supplement 1—source data 1
Raw images for western blot shown in Figure 2—figure supplement 1B (labelled).
- https://cdn.elifesciences.org/articles/98903/elife-98903-fig2-figsupp1-data1-v1.zip
-
Figure 2—figure supplement 1—source data 2
Raw images for western blot shown in Figure 2—figure supplement 1B.
- https://cdn.elifesciences.org/articles/98903/elife-98903-fig2-figsupp1-data2-v1.zip
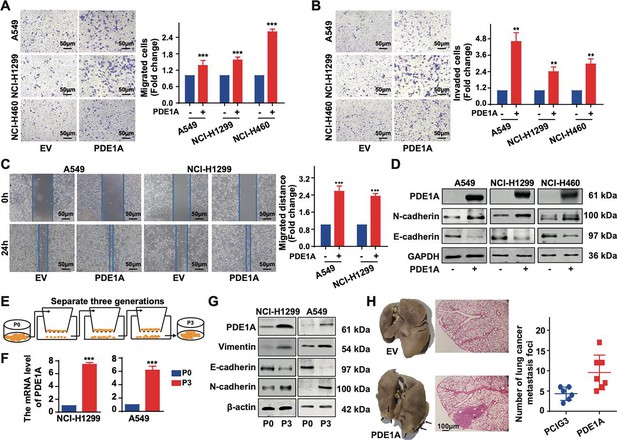
Phosphodiesterase 1A (PDE1A) promotes the metastasis and epithelial-mesenchymal transition (EMT) of non-small cell lung cancer (NSCLC) cells.
(A–B) NSCLC cells were transfected with PDE1A plasmid and empty vector for 24 hr. Cells were transferred to Transwell chambers without (A) or with (B) a Matrigel coating on the insert membrane, and the cell migrative and invasive abilities were determined, respectively, (n=3). (C) NSCLC cells were transfected with PDE1A plasmid and empty vector for 24 hr, and the wound healing assay was established in NSCLC cells, (n=3). (D) NSCLC cells were transfected with PDE1A plasmid and empty vector for 48 hr, and the expression of indicated proteins was detected. (E) The highly invasive NSCLC cells were separated using the Transwell chamber assay, and P3 cells were obtained from P0 cells after three generations. (F–G) The messenger RNA (mRNA) (F) and protein (G) levels of indicated genes were determined in P3 and P0 NSCLC cells. (H) The pulmonary metastatic nodules were stained using H&E and Bouin’s solution and counted in nude mice harboring NCI-H1299 cells transfected with PDE1A plasmid and empty vector, (n=6). **P < 0.01, ***P < 0.001.
-
Figure 3—source data 1
Raw images for western blots shown in Figure 3 (labelled).
- https://cdn.elifesciences.org/articles/98903/elife-98903-fig3-data1-v1.zip
-
Figure 3—source data 2
Raw images for western blots shown in Figure 3.
- https://cdn.elifesciences.org/articles/98903/elife-98903-fig3-data2-v1.zip
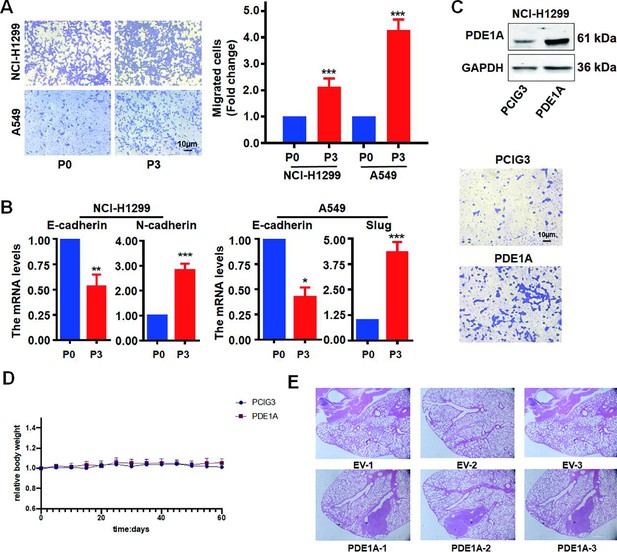
Phosphodiesterase 1A (PDE1A) promotes the metastasis of non-small cell lung cancer (NSCLC) cells.
(A) The migrative abilities of P3 and P0 NSCLC cells were determined, (n=3). (B) The messenger RNA (mRNA) levels of indicated genes were determined in P3 and P0 NSCLC cells using quantitative real‑time PCR (qRT-PCR), (n=3). (C) Before the NSCLC cells were injected into the nude mice, the migrative abilities of NSCLC cells were determined using Transwell assay. (D) The relative body weight of each group in the in vivo metastasis studies in Figure 3H, (n=6). (E) The lungs in nude mice were stained using H&E. **P < 0.01, ***P < 0.001.
-
Figure 3—figure supplement 1—source data 1
Raw images for western blots shown in Figure 3—figure supplement 1C (labelled).
- https://cdn.elifesciences.org/articles/98903/elife-98903-fig3-figsupp1-data1-v1.zip
-
Figure 3—figure supplement 1—source data 2
Raw images for western blots shown in Figure 3—figure supplement 1C.
- https://cdn.elifesciences.org/articles/98903/elife-98903-fig3-figsupp1-data2-v1.zip
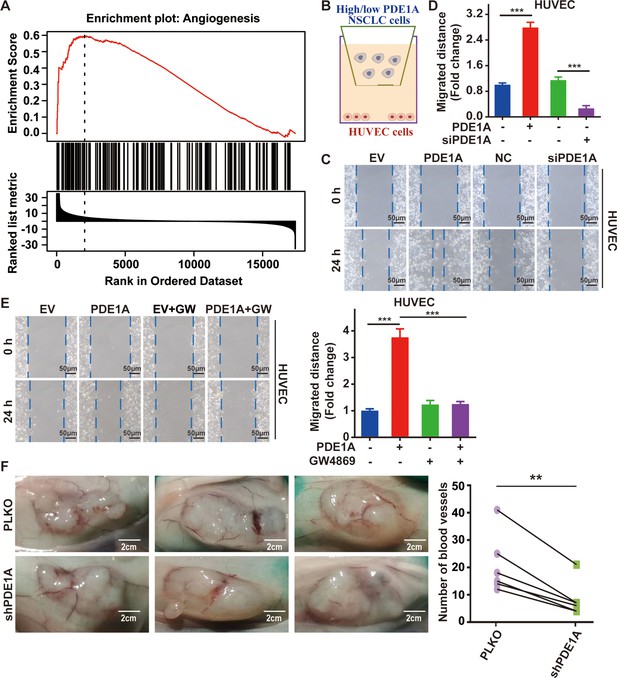
Non-small cell lung cancer (NSCLC) cells overexpressing phosphodiesterase 1A (PDE1A) promote angiogenesis in the tumor microenvironment (TME).
(A) It was collected from LinkedOmics. Statistical tests in LinkFinder include Pearson’s correlation coefficient, Spearman’s rank correlation, Student’s t-test, Wilcoxon test, analysis of variance, Kruskal-Wallis analysis, Fisher’s exact test, Chi-squared test, Jonckheere’s trend test, and Cox’s regression analysis. Multiple-test correction is performed using the Benjamini and Hochberg method to generate the false discovery rate. (B–D) NSCLC cells were transfected with empty vector/PDE1A overexpressing plasmid or control siRNA/siPDE1A for 48 hr, and then NSCLC cells were placed on the upper panel of Transwell with 0.4 μm insert, human umbilical vein endothelial cells (HUVECs) were placed on the lower panel of Transwell, and wound healing assay was performed to determine the migrative abilities of HUVECs, (n=3). (E) NSCLC cells with PDE1A overexpression were treated with 10 µM GW4869, and a wound healing assay was performed to determine the migrative abilities of HUVECs, (n=3). (F) NSCLC cells were transfected with empty vector or shPDE1A, then cells were transplanted into nude mice via subcutaneous injection, and the blood vessels were counted after 60 days, (n=6). **P < 0.01, ***P < 0.001.
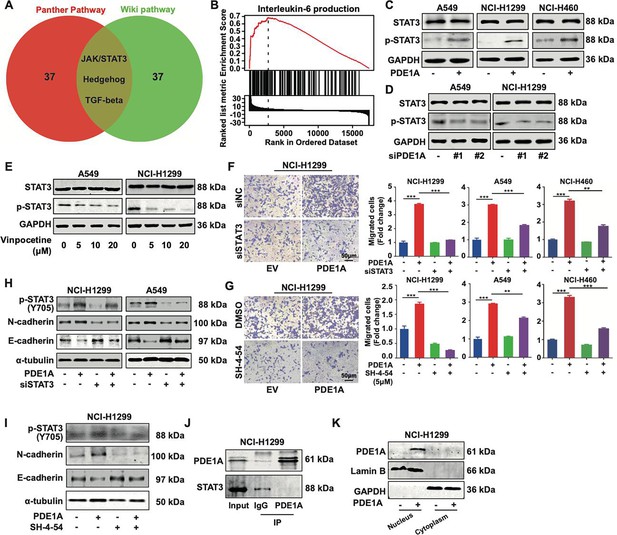
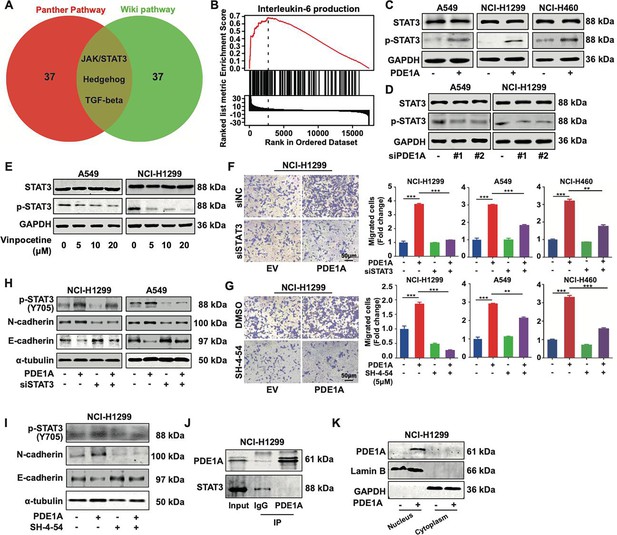
Phosphodiesterase 1A (PDE1A) promotes the metastasis of non-small cell lung cancer (NSCLC) cells via the STAT3 signaling pathway.
(A) A Venn diagram was generated using LinkedOmics, and overrepresentation enrichment analysis (ORA) was performed to analyze the molecular pathway regulated by PDE1A in NSCLC. Sample cohort: TCGA_NSCLC; Institute: UNC; Data type: RNAseq; Platform: HiSeq RNA; Attribute: PDE1A; Statistical methods: Spearman’s correlation test; Patients: 515; Tools: ORA; Gene Ontology analysis: WikiPathways and PANTHER Pathway; Select rank criteria: FDR; Select sign: Positively correlated; Significance level: 0.05; TOP40 was selected to generate the Venn diagram. (B) Gene set enrichment analysis (GSEA) was performed to analyze the biological process of PDE1A in NSCLC. (C) NSCLC cells were transfected with PDE1A plasmid and empty vector for 48 hr, and the expression of indicated proteins was detected. (D) NSCLC cells were transfected with control siRNA and PDE1A siRNA for 48 hr, and the expression of indicated proteins was detected. (E) NSCLC cells were treated with DMSO or vinpocetine (5, 10, 20 µM) for 24 hr, and the expression of indicated proteins was determined. (F) NSCLC cells overexpressing PDE1A were transfected with control siRNA and STAT3 siRNA for 48 hr, and the migrative abilities of NSCLC cells were determined by Transwell assay, (n=3). (G) NSCLC cells overexpressing PDE1A were treated with STAT3 inhibitor SH-4–54 (5 µM) for 24 hr. The migrative abilities of NSCLC cells were determined by Transwell assay, (n=3). (H) NSCLC cells overexpressing PDE1A were transfected with control siRNA and STAT3 siRNA for 48 hr, and the expression of indicated protein was detected by western blot. (I) NSCLC cells overexpressing PDE1A were treated with STAT3 inhibitor SH-4–54 (5 µM) for 24 hr, and the expression of indicated protein was detected by western blot. (J) The interaction between PDE1A and STAT3 was determined by immunoprecipitation. (K) NCI-H1299 cells were transfected with empty vector and PDE1A overexpressing plasmid for 48 hr, and the contribution of PDE1A in the cytoplasm and nucleus was determined.**P < 0.01, ***P < 0.001.
-
Figure 5—source data 1
Raw images for western blots shown in Figure 5 (labelled).
- https://cdn.elifesciences.org/articles/98903/elife-98903-fig5-data1-v1.zip
-
Figure 5—source data 2
Raw images for western blots shown in Figure 5.
- https://cdn.elifesciences.org/articles/98903/elife-98903-fig5-data2-v1.zip
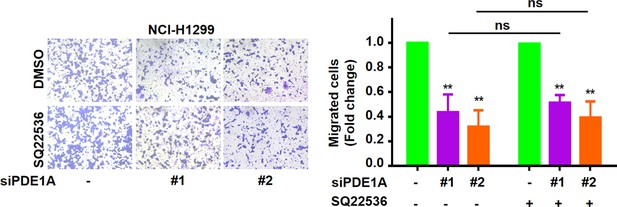
Phosphodiesterase 1A (PDE1A) promotes the metastatic potential in a cyclic adenosine monophosphate (cAMP)-independent manner.
Non-small cell lung cancer (NSCLC) cells were transfected with control siRNA and PDE1A siRNA. After 24 hr, cells were collected for Transwell assay. Cells were treated with DMSO or 5 µM SQ22536 for 24 hr, and the migrative abilities of NSCLC cells were determined, (n=3). ns, not significant, **P < 0.01.
-
Figure 5—figure supplement 1—source data 1
Raw images for western blots related to Figure 5—figure supplement 1 (labelled).
- https://cdn.elifesciences.org/articles/98903/elife-98903-fig5-figsupp1-data1-v1.zip
-
Figure 5—figure supplement 1—source data 2
Raw images for western blots related to Figure 5—figure supplement 1.
- https://cdn.elifesciences.org/articles/98903/elife-98903-fig5-figsupp1-data2-v1.zip
PDE1A physically interacts with YT521-B homology domain family member 2 (YTHDF2) and promotes the metastasis of non-small cell lung cancer (NSCLC) cells.
(A) Venn diagram showing the overlap among PDE1A-interacting proteins (it was collected from mass spectrometry analysis in NSCLC cells), STAT3-coexpressed genes (collected from gene correlation using UALCAN), upregulated proteins in NSCLC compared with normal tissues (analyzed by UALCAN based on CPTAC database), and upregulated genes in NSCLC compared with normal tissues (analyzed by UALCAN based on TCGA database). Pearson’s correlation analysis of UALCAN was used to evaluate gene correlation analyses, and Welch’s t-test was estimated to detect the significance of differences in expression levels between two groups. (B) GO enrichment analysis of PDE1A-interacting genes. (C) Immunoprecipitation followed by silver staining was performed to identify protein and protein interaction using A549 cell lysate with the anti-PDE1A antibody. (D) Immunoprecipitation was used to confirm protein and protein interaction in NCI-H1299 cells. (E) NSCLC cells overexpressing PDE1A were transfected with control siRNA and YTHDF2 siRNA for 48 hr, and the migrative abilities of NSCLC cells were determined by Transwell assay, (n=3). (F) NSCLC cells overexpressing PDE1A were transfected with control siRNA and YTHDF2 siRNA for 48 hr, and the expression of indicated protein was detected by western blot. ***P < 0.001.
-
Figure 6—source data 1
Raw images for western blots shown in Figure 6 (labelled).
- https://cdn.elifesciences.org/articles/98903/elife-98903-fig6-data1-v1.zip
-
Figure 6—source data 2
Raw images for western blots shown in Figure 6.
- https://cdn.elifesciences.org/articles/98903/elife-98903-fig6-data2-v1.zip
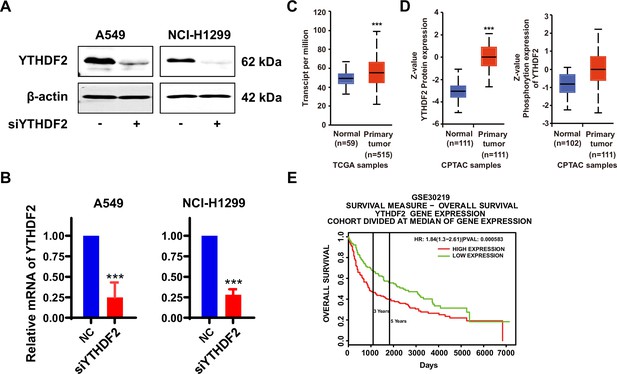
YT521-B homology domain family member 2 (YTHDF2) predicts poor outcomes for lung cancer patients.
(A–B) Non-small cell lung cancer (NSCLC) cells were transfected with control siRNA and YTHDF2 siRNA for 48 hr, and the knockdown efficiency of YTHDF2 was confirmed by western blot (A) and quantitative real‑time PCR (qRT-PCR) (B), (n=3). (C–D) The messenger RNA (mRNA) and protein levels of phosphodiesterase 1A (PDE1A) in NSCLC and normal lung tissues are shown. It was collected from UALCAN and shown. Gene: YTHDF2; TCGA dataset: Lung adenocarcinoma (C); CPTAC dataset: Lung adenocarcinoma (D). (E) The prognostic value of YTHDF2 in NSCLC patients was identified using the PROGgeneV2 online tool (https://www.progtools.net/gene). ***P < 0.001.
-
Figure 6—figure supplement 1—source data 1
Raw images for western blots shown in Figure 6—figure supplement 1A (labelled).
- https://cdn.elifesciences.org/articles/98903/elife-98903-fig6-figsupp1-data1-v1.pdf
-
Figure 6—figure supplement 1—source data 2
Raw images for western blots shown in Figure 6—figure supplement 1A.
- https://cdn.elifesciences.org/articles/98903/elife-98903-fig6-figsupp1-data2-v1.zip
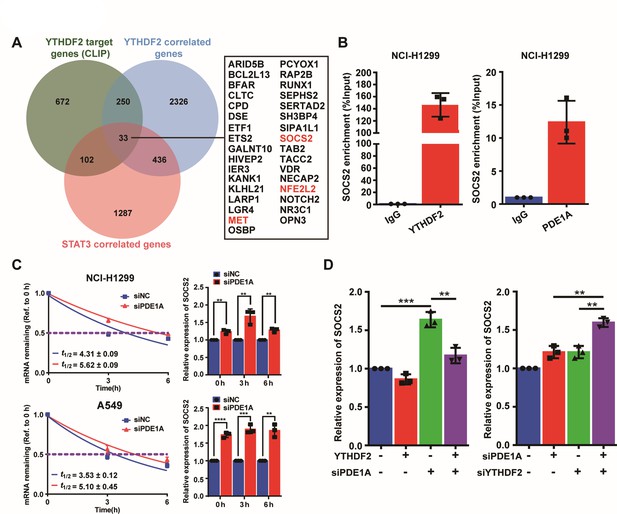
Phosphodiesterase 1A (PDE1A) interacts with YT521-B homology domain family member 2 (YTHDF2) to regulate SOCS2/STAT3 signaling pathway.
(A) YTHDF2-RNA complexes were identified by LC-MS/MS and collected from reference; YTHDF2 correlated genes were collected from TNMplot (https://tnmplot.com/analysis/), Gene: YTHDF2, Gene vs. all genes correlation: Genechip data, Tissue: Lung; STAT3 correlated genes were collected from cBioPortal (http://www.cbioportal.org/), Lung cancer (SMC, cancer research 2016); n=22; Gene: STAT3. The interaction between YTHDF2 protein and the messenger RNA (mRNA) of 33 overlapping genes was predicted by RNA-Protein Interaction Prediction (http://pridb.gdcb.iastate.edu/RPISeq/index.html), and the values of RF classifier and SVM classifier above 0.5 were considered positive. Comparison of the normal and the tumorous samples was performed by the Mann-Whitney U test, and normal, tumorous, and metastatic tissue gene comparison can be analyzed using the Kruskal-Wallis test. (B) The interactions between protein and mRNAs were verified by RNA binding protein immunoprecipitation (RIP) experiments. (C) Non-small cell lung cancer (NSCLC) cells were transfected with control siRNA and siPDE1A for 48 hr, and the stability of mRNA was determined by quantitative real‑time PCR (qRT-PCR), (n=3). (D) NSCLC cells were transfected with control siRNA and siPDE1A for 48 hr, and the expression of SOCS2 mRNA was determined by qRT-PCR, (n=3). **P < 0.01, ***P < 0.001.
Additional files
-
Supplementary file 1
The siRNA sequence used for knocking down the indicated genes.
- https://cdn.elifesciences.org/articles/98903/elife-98903-supp1-v1.xlsx
-
Supplementary file 2
Primers sequences for detecting the expression of the indicated genes.
- https://cdn.elifesciences.org/articles/98903/elife-98903-supp2-v1.xlsx
-
Supplementary file 3
Identification of PDE1A protein interactions by mass spectrometry.
- https://cdn.elifesciences.org/articles/98903/elife-98903-supp3-v1.pdf
-
Supplementary file 4
The potential target genes of YTHDF2 in non-small cell lung cancer (NSCLC) cells.
- https://cdn.elifesciences.org/articles/98903/elife-98903-supp4-v1.xlsx
-
Supplementary file 5
The potential target genes of YTHDF2 in non-small cell lung cancer (NSCLC) cells.
- https://cdn.elifesciences.org/articles/98903/elife-98903-supp5-v1.xlsx
-
MDAR checklist
- https://cdn.elifesciences.org/articles/98903/elife-98903-mdarchecklist1-v1.docx





