PRR adjuvants restrain high stability peptides presentation on APCs
Figures

Immunodominant T-cell epitopes in different adjuvants vaccination mice.
Spleens were collected from mice on day 10 post-vaccination with the antigen UreB incorporated with adjuvants CpG, MDP, and MPLA; cultured in vitro; and stimulated with a panel of overlapping UreB 18 mer peptides to assess the responsiveness of CD4+ T-cells for interferon-γ (IFN-γ) using ICS. The percentages of CD4+ T-cells secreting IFN-γ against each peptide were determined using flow cytometry. Locations of the dominant peptides in different groups are indicated. The results are representative of three independent experiments.
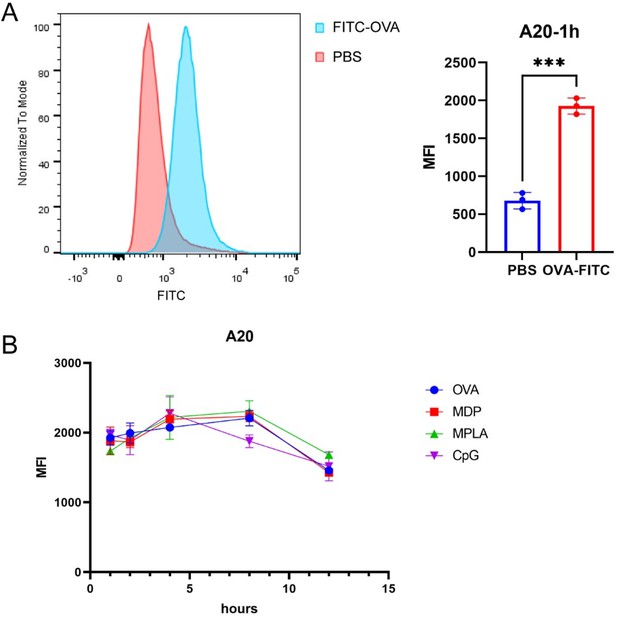
MHC-II peptidome and proteome measurements in adjuvant-treated antigen-presenting cells (APCs).
A20 cells were treated with CpG ODN, MDP, or MPLA incorporated with Helicobacter pylori antigens for 12 hr. Most cells (108) were lysed for immunopeptidomics and the remaining cells (107) were used for proteomics. (A) Experimental flow chart. (B) Number of MHC peptides identified in the different adjuvant-treated groups (n=3, biological replicates). The numbers indicate mean values. (C) Length distribution of MHC peptides in different adjuvant-treated groups. (D) Sequence motifs of the MHC peptides identified in the adjuvant-treated groups. (E) Binding heatmaps of all eluted MHC peptides between 9–22 mer in adjuvant-treated groups were predicted and assigned to alleles using NetMHCIIpan. ns: not significantly different (p>0.05).
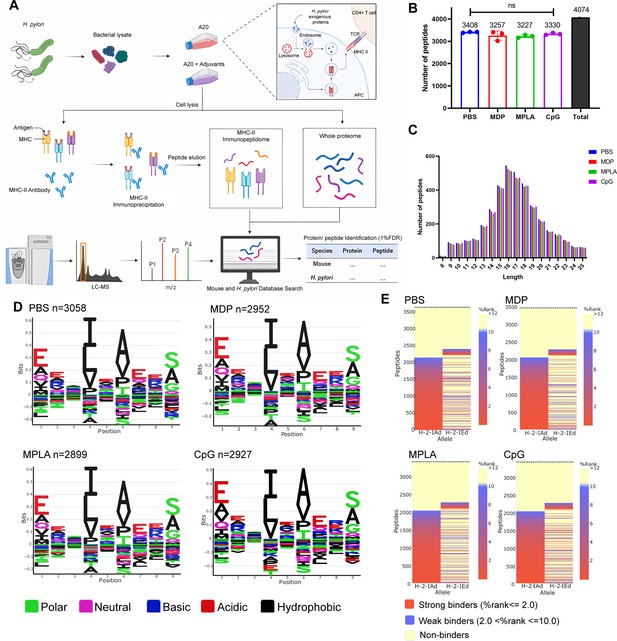
Peptide logos and MHC-II binding are assigned to individual alleles.
Logo plots of the identified MHC peptides for individual alleles H2-IA and H2-IE were analyzed. The fractions of MHC peptides between 9–22 mer binding to each allele are shown.
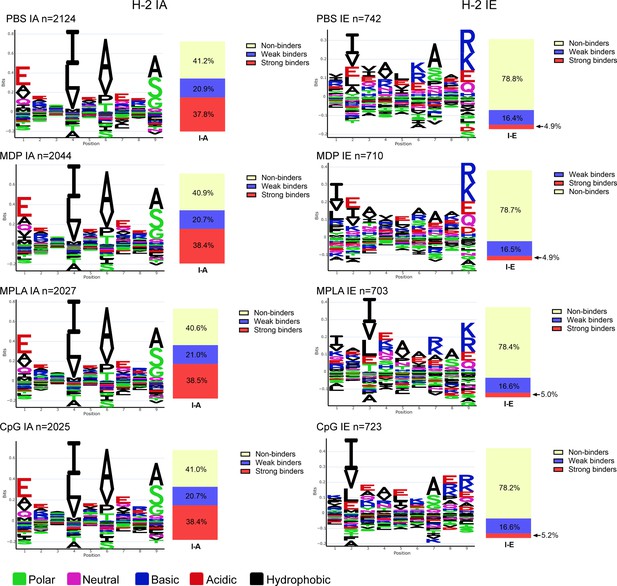
Immunopeptidomics of J774A.1 cell line and surface marker detection.
(A) 5×108 J774A.1 cells and 108 A20 cells were used for immunopeptidomics. The number of MHC peptides was compared. Less than 350 MHC peptides in J77A.1 cells and more than 5500 MHC peptides in A20 cells were observed at a peptide spectrum match (PSM) level of <1.0% false discovery rate (FDR). (B) J774.1 and A20 cell surface markers were stained with specific antibodies or corresponding isotype controls. Then, expression levels were compared between J774.1 and A20 cells (n=3, biological replicates). MFI, mean fluorescence intensity. ****p<0.0001.
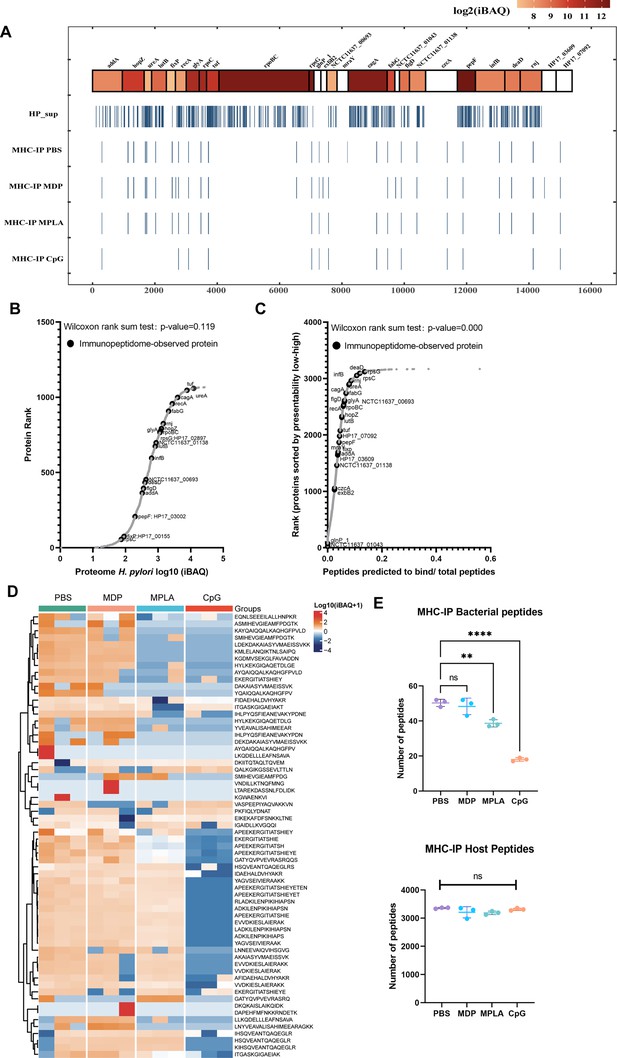
Profiling exogenous MHC-II peptides in adjuvant-treated antigen-presenting cells (APCs).
(A) Peptide locations across the Helicobacter pylori genome from MHC-II immunopeptidomes. (B) Rank plot of each protein abundance detected in the whole-proteome of bacterial ultrasonic supernatant antigens. Proteins identified in immunopeptidomes are annotated with their respective gene names. (C) MHC-II presentation potential of bacterial proteins. All reported H. pylori proteins were ranked according to the ratio between the number of peptides predicted to be presented by MHC-II alleles (rank ≤2) and the total number of 13- to 17-mer. Proteins identified in immunopeptidomes are annotated with their respective gene names. (D) Heatmap of exogenous MHC peptides from different adjuvant groups. The identified sequences are shown. (E) Numbers of MHC peptides derived from bacteria and hosts were compared among different adjuvant groups. n=3. **p<0.01, ****p<0.0001.
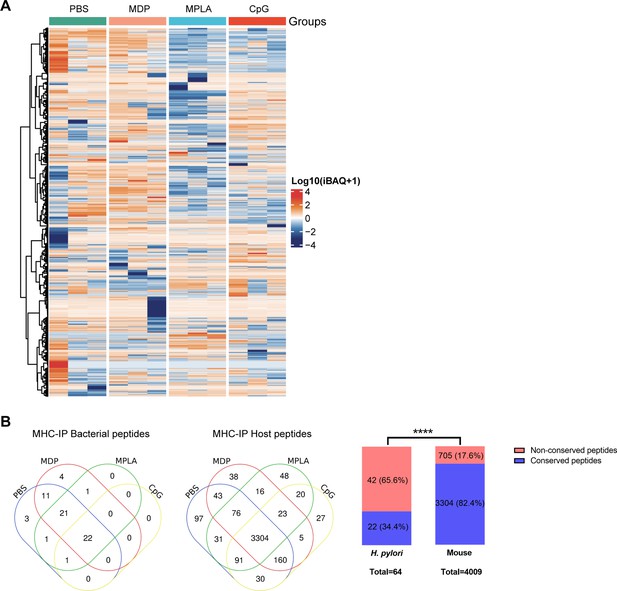
Profiling peptides from MHC-II immunopeptidomes.
(A) Heatmap showing the host MHC peptides from the MHC-II immunopeptidome. (B) Venn diagrams showing the distribution of bacterial and host MHC peptides in different adjuvant groups. Fractions of bacterial and host-conserved and non-conserved MHC peptides were analyzed. The number and percentage of peptides are indicated. ****p<0.0001.
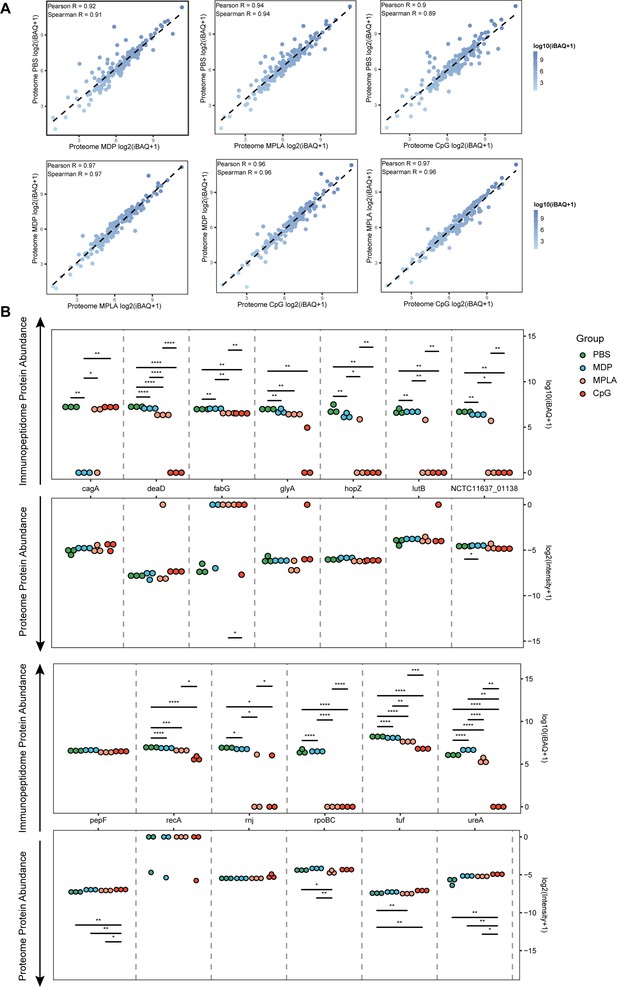
Antigen phagocytosis of antigen-presenting cells (APCs) treated with different adjuvants.
(A) Comparison of bacterial protein abundance in APCs 12 hr post-adjuvant stimulation from whole proteomes. (B) Abundances of bacterial proteins from immunopeptidome and proteome were compared among adjuvant groups. *p<0.05, **p<0.01, ***p<0.001, ****p<0.0001.
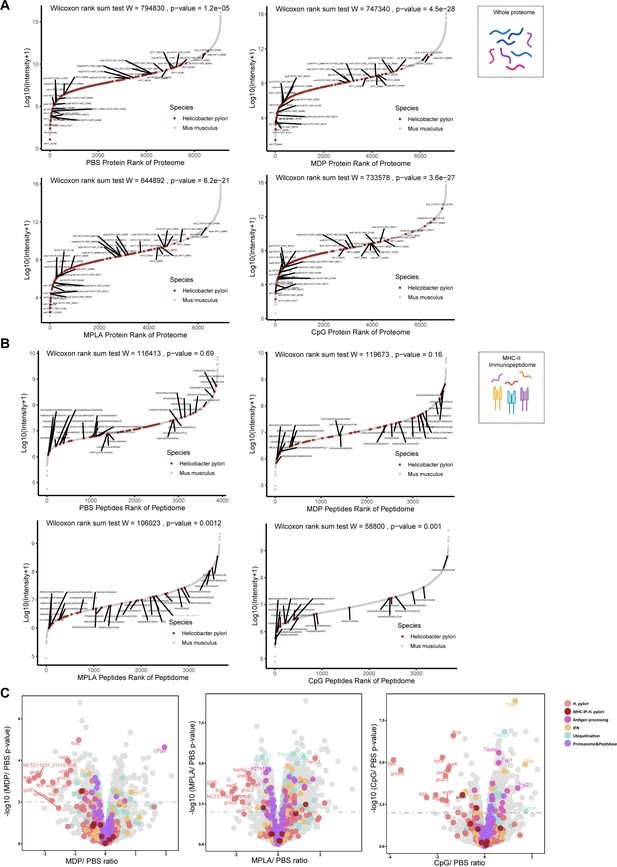
The effects of adjuvants on antigen presentation.
(A) Rank plot of host and bacterial protein abundances in the whole-proteome and (B) MHC–peptide abundances from immunopeptidomes of different adjuvant groups. Bacterial proteins are marked with red, and some of them are annotated with their respective gene names. Bacterial MHC peptides are annotated with their respective amino acid sequence. (C) Volcano plots comparing protein levels between PBS- and adjuvant-treated groups in the whole-proteome. Proteins involved in antigen processing, ubiquitination, proteasome, and peptidase, and interferon (IFN) pathways are colored accordingly. Above the dashed line (p<0.01) means significant.
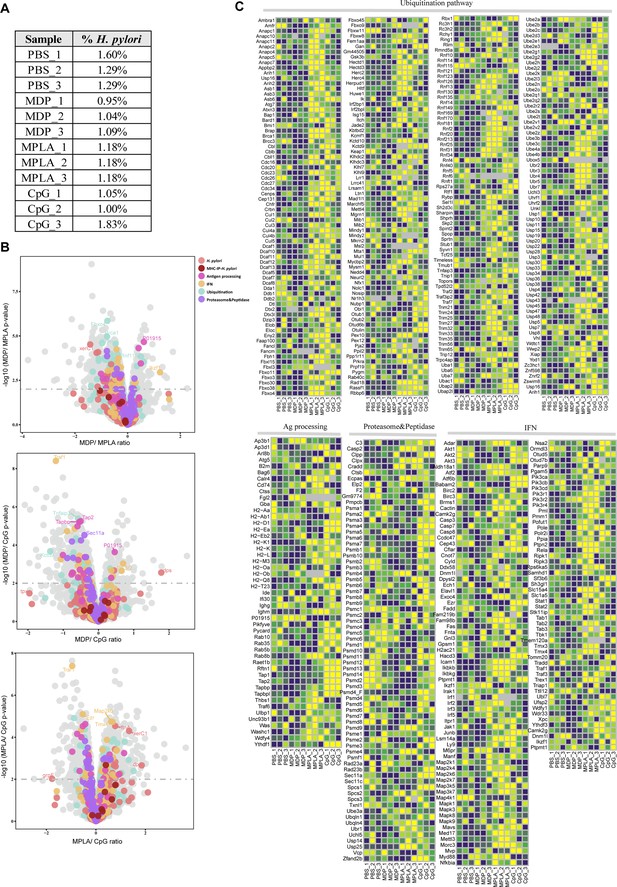
Bacterial proteins and host antigen presentation proteins in the whole-proteome of different adjuvant groups.
(A) Percentage of bacterial protein abundances in the total whole proteomes. (B) Volcano plots comparing protein levels among adjuvant-treated groups (the dashed line: p<0.01). (C) Abundances of proteins (log10 protein iBAQ) involved in antigen processing and presentation in the whole-proteome analysis.
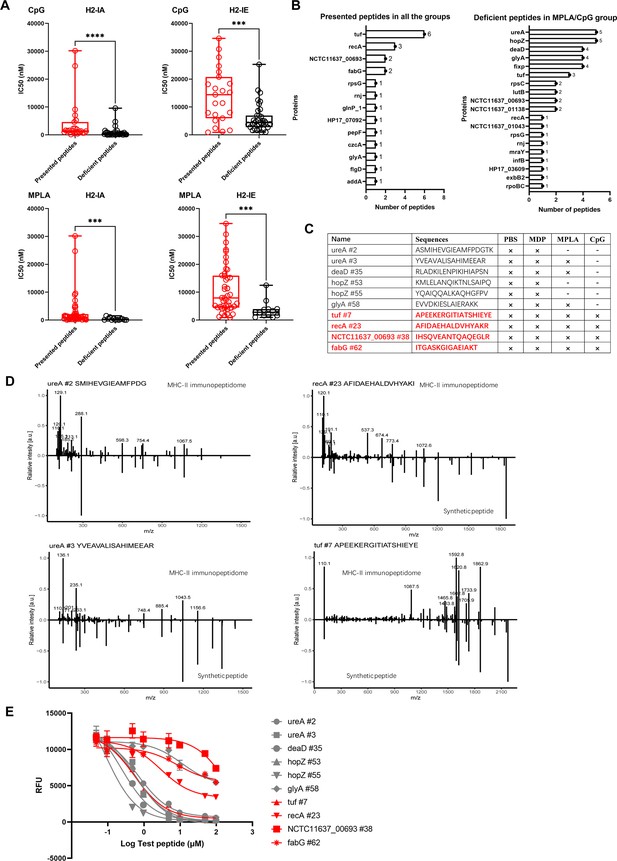
Binding affinity of MS-detected peptides was determined.
(A) IC50 of the presented and deficient peptides post-adjuvant stimulation from immunopeptidome binding to H2-IA and H2-IE were predicted by the NN align method using the IEDB website. The data of peptides from adjuvants MPLA and CpG are shown. High IC50 means low binding stability. Boxes show quartiles, bars indicate medians, and whiskers show distributions. (B) Distribution of proteins corresponding to bacterial MHC peptides from immunopeptidome. The numbers of peptides identified by MS for each protein are indicated. (C) Information of 10 synthetic peptides from Top4 presented and deficient proteins. ×: Presence of peptides in the corresponding group. -: Peptides missing in the corresponding group. (D) Mirror plots with fragment ion mass spectra to confirm the sequences of MHC peptides from immunopeptidome. Positive y-axis, MHC-II IP sequences; negative y-axis, synthetic peptides. (E) Competitive binding curve of synthetic peptides for MHC-II H2-IA allele. n=3. The binding curves of peptides presented in adjuvant groups are marked with red.
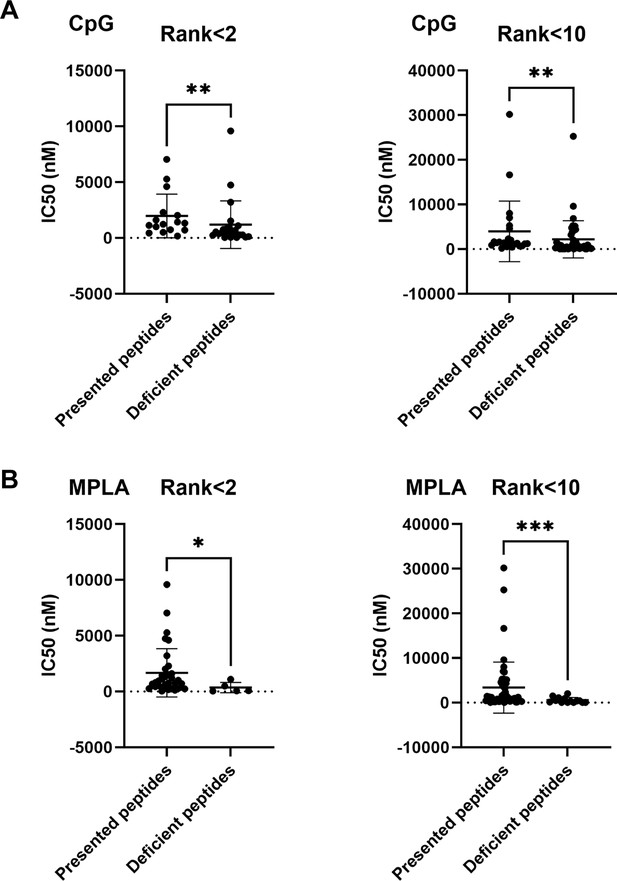
Binding affinities of bacterial MHC peptides were assessed at cutoffs of percentile rank <2 (Strong Binders) or <10 (Weak Binders).
Peptides binding to MHC-II alleles were screened at cutoffs of percentile rank <2 (Strong Binders) or <10 (Weak Binders) by the NetNHCIIpan_el 4.1 method using the IEBD website. The IC50 of these peptides was predicted and compared using the NN alignment method. IC50 of the presented and deficient peptides after (A) CpG and (B) MPLA stimulation at cutoffs of percentile ranks <2 and <10 were analyzed. *p<0.05, **p<0.01, ***p<0.001.

T-cell responses induced by MS-detected peptides with different binding stability were analyzed.
Five BALB/c mice were immunized with a pool of 10 synthetic peptides. On days 10 and 28 post-immunization, the splenocytes were isolated and stimulated with individual peptides for interferon-γ (IFN-γ) Elisopt assay. (A) Experimental flow chart. (B, C) Elispot results on days 10 and 28. OVA peptide and non-stimulated wells were used as negative controls. PMA stimulation was used as the positive control. Responses to the peptides in the adjuvant groups are marked in red. The dashed line represents the 3x median of the OVA peptide used as the threshold for a positive response. Boxes show quartiles, bars indicate medians, and whiskers show distributions. Elispot images of positive responses from one immunized mouse are shown. Numbers indicate spot counts. (D) Epitope-specific CD4+ T-cells from immunized mice spleens were expanded in vitro and the IFN-γ-producing CD4+ T-cells were assessed using the peptides pool (left). The present peptide recA #23 and deficient peptides ureA #2 and ureA #3 after adjuvant treatment were titrated to restimulate the expanded cells (right, n=3). The IFN-γ responses of CD4+ T-cells were detected by FACS. The responses induced by the indicated peptides at 50 μM were considered 100%. All other responses were evaluated based on their relative strengths.
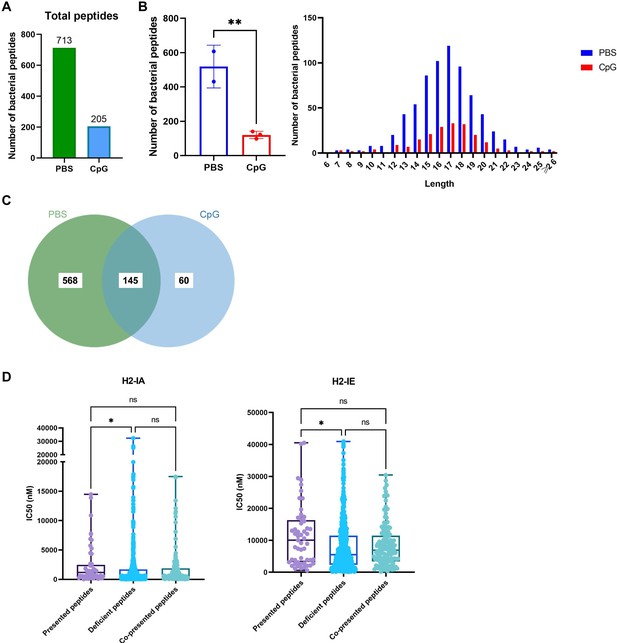
MHC-II peptidome measurements in adjuvant-treated APCs pulsed with Pseudomonas aeruginosa antigens.
(A) Total number of bacterial peptides identified in the PBS- and CpG-treated groups. (B) The number and length distribution of bacterial peptides in different groups were compared. (C) Venn diagrams showing the distribution of bacterial peptides in different groups. (D) IC50 of the presented, deficient, and co-presented peptides post-adjuvant stimulation from immunopeptidome binding to H2-IA and H2-IE were predicted using the IEDB website. High IC50 means low binding stability. *p<0.05, **p<0.01.
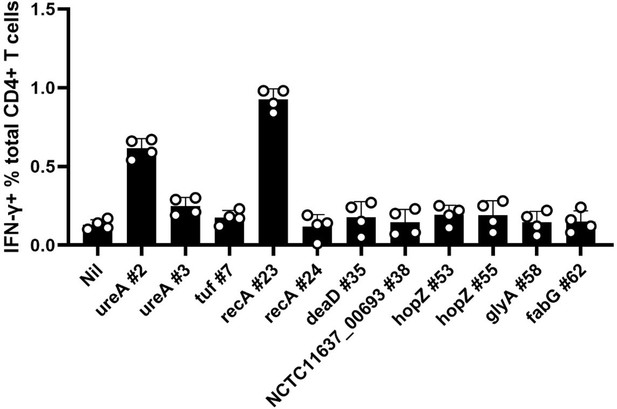
The expanded CD4+T cells from peptides immunized mice were screened for their response to the peptides in an ICS assay.
Tables
| Reagent type (species) or resource | Designation | Source or reference | Identifiers | Additional information |
|---|---|---|---|---|
| Strain, strain background (Helicobacter pylori) | NCTC 11637; ATCC 43504 | ATCC | Cat#: 43504 | |
| Cell line (Mus musculus) | A20 (B cell lymphoma line, BALB/cAnN mouse) | ATCC | Cat#: TIB-208 | |
| Cell line (Mus musculus) | J774A.1 (monocyte/ macrophage cell line, BALB/cN mouse) | ATCC | Cat#: TIB-67 | |
| Biological sample (BALB/c Mouse, female) | Primary splenic cells | VITALSTAR, China | Freshly isolated from mouse | |
| Antibody | Anti-mouse CD45-FITC (Rat monoclonal) | Biolegend | Cat#: 103108 | FACS (0.5 ul per test) |
| Antibody | Anti-mouse CD3-PE-Cy7 (Armenian Hamster monoclonal) | Biolegend | Cat#: 100320 | FACS (2 ul per test) |
| Antibody | Anti-mouse IFN-γ- PE/Dazzle594 (Rat monoclonal) | Biolegend | Cat#: 505846 | FACS (0.3 ul per test) |
| Antibody | Anti-mouse MHC-II-APC (Rat monoclonal) | Biolegend | Cat#: 107614 | FACS (1 ul per test) |
| Antibody | Anti-mouse CD4-APC (Rat monoclonal) | Biolegend | Cat#: 100412 | FACS (1 ul per test) |
| Antibody | Anti-mouse CD86-PE (Rat monoclonal) | Biolegend | Cat#: 105007 | FACS (5 ul per test) |
| Antibody | Anti-mouse CD80- FITC (Armenian Hamster monoclonal) | Biolegend | Cat#: 104706 | FACS (2 ul per test) |
| Antibody | Anti-mouse CD45- APC-Cy7 (Rat monoclonal) | Biolegend | Cat#: 147718 | FACS (1 ul per test) |
| Antibody | Anti-mouse CD16/32 TruStain FcX PLUS (Rat monoclonal) | Biolegend | Cat#: 156604 | FACS (1 ul per test) |
| Antibody | Anti-Mouse H2-IAd/IEd antibody (Rat monoclonal) | BioXcell | Cat#: BE00108 | IP (2 mg per sample) |
| Antibody | Anti- Mouse MHC-II (I-A/I-E) antibody (Rat monoclonal) | Thermo Fisher Scientific | Cat#: 14-5321-82 | MHC binding assay (10 μg/ ml) |
| Peptide, recombinant protein | Recombinant Murine IL-2 | PeproTech | Cat#: 212–12 | |
| Peptide, recombinant protein | Urease B subunit | This paper | Recombinant protein | Purified (purity >95%) by ourselves. |
| Peptide, recombinant protein | All peptides used in this paper | China Peptides | peptide | Customized (sequences were shown in the paper) |
| Peptide, recombinant protein | Purified mouse MHC-II | Proimmune | Customized | |
| Commercial assay or kit | Mouse IFN-γ precoated ELISpot kit | Dakewe | Cat#: 2210003 | |
| Commercial assay or kit | Cyto-Fast Fix/Perm Buffer Set | Biolegend | Cat#: 426803 | |
| Chemical compound, drug | MPLA-SM | Invivogen | Cat#: tlrl-mpla2 | |
| Chemical compound, drug | MDP | Invivogen | Cat#: tlrl-mdp | |
| Chemical compound, drug | CPG ODN | Invivogen | Cat#: tlrl-1826 | |
| Chemical compound, drug | Brefeldin A Solution | Biolegend | Cat#: 420601 | |
| Chemical compound, drug | CNBr-activated Sepharose | Cytivia | Cat#: 17-0430-01 | |
| Chemical compound, drug | CHAPS | Millipore | Cat#: 1116620001 | |
| Chemical compound, drug | Europium-Streptavidin | Abcam | Cat#: ab270228 | |
| Chemical compound, drug | Complete Freund’s Adjuvant, CFA | Sigma-Aldrich | Cat#: F5881-10ML | |
| Software, algorithm | SPSS | SPSS | RRID:SCR_002865 | |
| Software, algorithm | GraphPad Prism | GraphPad Prism | RRID:SCR_002798 | |
| Software, algorithm | MHC-II binding prediction | IEDB | RRID:SCR_006604 | https://www.iedb.org/ |
| Software, algorithm | Prediction of peptide-MHC-II binding motifs | MhcVizPipe (CaronLab, 2021) | https://github.com/CaronLab/MhcVizPipe/ | |
| Other | Aseptic rabbit blood | Sbjbio | Cat#: SBJ-ST-RAB002 | For making H. pylori culture plates |
Additional files
-
Supplementary file 1
Whole-proteome data, related to Figure 5 and Figure 5—figure supplement 1.
The whole proteome data of A20 cells treated with different adjuvants at 12 hr. The ‘1 a’ sheet contains the KEGG and GO annotations of proteins from antigen processing, peptidase function, ubiquitination pathway, interferon (IFN) signaling, and H. pylori. The ‘1b’ sheet contains the iBAQ values of each protein in different adjuvant groups.
- https://cdn.elifesciences.org/articles/99173/elife-99173-supp1-v1.xlsx
-
MDAR checklist
- https://cdn.elifesciences.org/articles/99173/elife-99173-mdarchecklist1-v1.pdf