A lytic transglycosylase connects bacterial focal adhesion complexes to the peptidoglycan cell wall
Figures
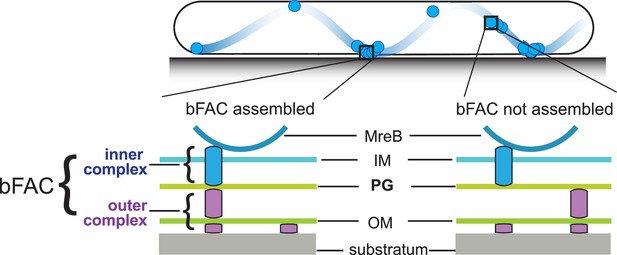
Stationary bacterial focal adhesion complexes (bFACs) drive M. xanthus gliding.
Motors carrying incomplete gliding complexes either diffuse or move rapidly along helical paths but do not generate propulsion. Motors stall and become nearly static relative to the substrate when they assemble into complete bFACs with other motor-associated proteins at the ventral side of the cell. Stalled motors push MreB and bFACs in opposite directions and thus exert force against outer membrane adhesins. Overall, as motors transport bFACs toward lagging cell poles, cells move forward but bFACs remain static relative to the substrate. IM, inner membrane; OM, outer membrane.

AgmT, a putative lytic transglycosylase, is required for M. xanthus gliding.
(A) AgmT shows significant similarity to a widely conserved peptidoglycan (PG) transglycosylase in the YceG/MltG family. The conserved glutamine residue is marked by an asterisk. M_xant, M. xanthus; E_coli, E. coli; M_tube, Mycobacterium tuberculosis; S_coel, Streptomyces coelicolor; A_calc, Acinetobacter calcoaceticus; X_albi, Xanthomonas albilineans; N_meni, Neisseria menningitidis; R_prow, Rickettsia prowazekii; T_aqua, Thermus aquaticus. (B) AgmT is required for M. xanthus gliding. Colony edges were imaged after incubating cells on 1.5% agar surfaces for 24 hr. To eliminate S-motility, we further knocked out the pilA gene that encodes pilin for type IV pilus. Cells that move by gliding are able to move away from colony edges (as pointed by the arrows in the inset). Deleting agmT or disabling the active site of AgmT (AgmTEAEA) abolish gliding but fusing an PAmCherry (PAmCh) to its C-terminus does not. Heterologous Expression of E. coli MltG (MltGEc) restores gliding of agmT cells but not the cells that express AgmTEAEA. While cells lacking AgmT moved slower (C) and less persistently (D, measured by the distances cells traveled before pauses and reversals), the expression of MltGEc restores both the velocity and persistency of gliding in the agmT cells. Data were pooled from three biological replicates and p values were calculated using a one-way analysis of variance (ANOVA) test between two unweighted, independent samples. Boxes indicate the 25th to 75th percentiles and bars the median. The total number of cells analyzed is shown on top of each plot. *p < 0.001. Growth curves of agmT-related mutants can be found in Figure 2—figure supplement 1. Other putative lytic transglycosylases (LTGs) do not affect gliding motility significantly, which is shown in Figure 2—figure supplement 2.
-
Figure 2—source data 1
Gliding velocity data for Figure 2C.
- https://cdn.elifesciences.org/articles/99273/elife-99273-fig2-data1-v1.xlsx
-
Figure 2—source data 2
Gliding persistency data for Figure 2D.
- https://cdn.elifesciences.org/articles/99273/elife-99273-fig2-data2-v1.xlsx
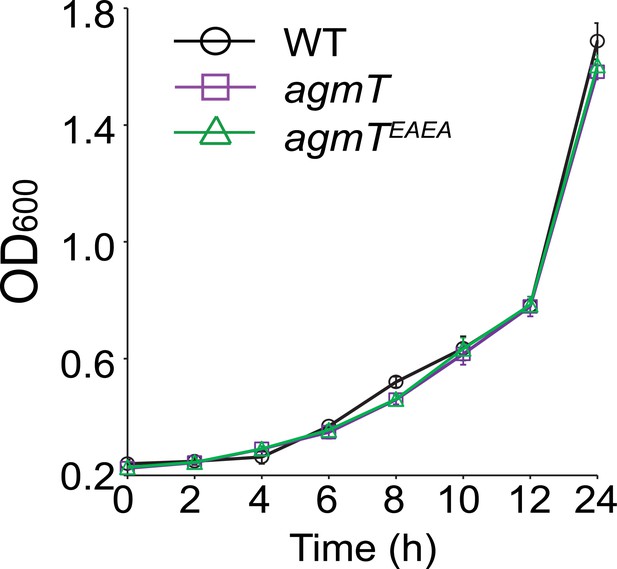
AgmT does not regulate growth.
Data are presented as mean ± standard deviation (SD) from three technical replicates.
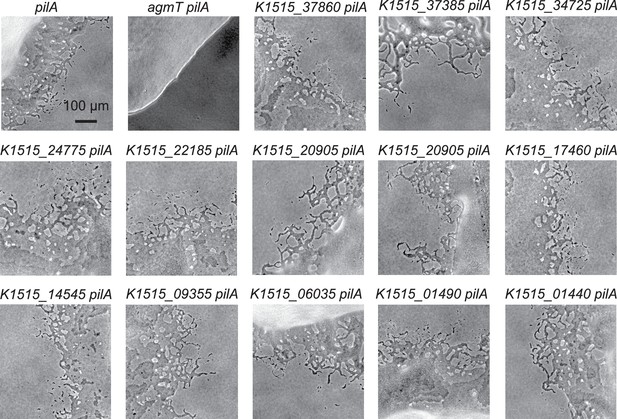
Other putative lytic transglycosylases (LTGs) are not required for M. xanthus gliding motility.
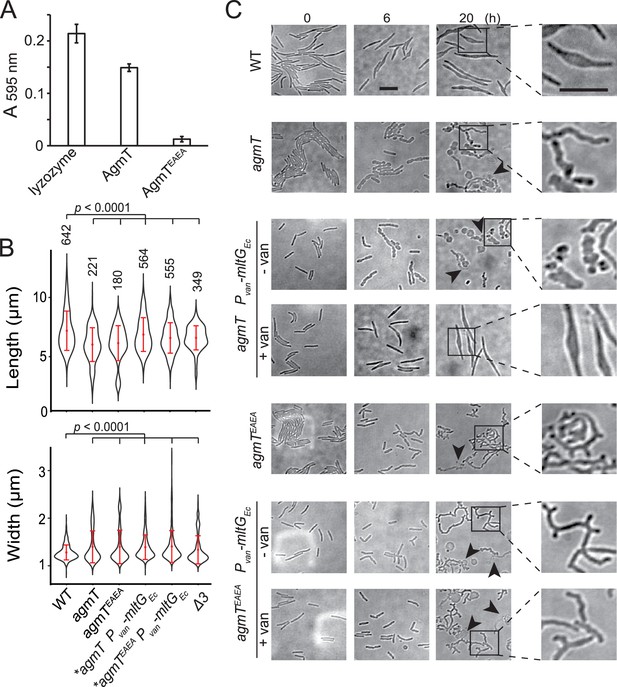
AgmT regulates cell morphology and integrity under antibiotic stress.
(A) Purified AgmT solubilizes dye-labeled peptidoglycan (PG) sacculi, but AgmTEAEA does not. Lysozyme serves as a positive control. Absorption at 595 nm was measured after 18 hr incubation at 25°C. Data are presented as mean values ± standard deviation (SD) from three technical replicates. (B) AgmT regulates cell morphology. Compared to wild-type cells, cells that lack AgmT and express AgmTEAEA are significantly shorter and wider. A previously reported mutant that lacks all three class A penicillin-binding proteins (Δ3) displays similarly shortened and widened morphology but is still motile by gliding (Figure 3—figure supplement 1). Heterologous expression of E. coli MltG partially restores cell length but not cell width in agmT and agmTEAEA backgrounds. Asterisks, 200 µM sodium vanillate added. Data were pooled from two biological replicates and p values were calculated using a one-way ANOVA test between two unweighted, independent samples. Whiskers indicate the 25th to 75th percentiles and red dots the median. The total number of cells analyzed is shown on top of each plot. (C) AgmT regulates cell morphology and integrity under mecillinam stress (100 μg/ml). Expressing E. coli MltG by a vanillate-inducible promoter (Pvan) restores resistance against mecillinam in agmT cells but not in the cells that express AgmTEAEA. van, 200 µM sodium vanillate. Arrows point to newly lysed cells. Scale bars, 5 µm.
-
Figure 3—source data 1
Absorption at 595 nm in the Remazol brilliant blue (RBB) assay for Figure 3C.
- https://cdn.elifesciences.org/articles/99273/elife-99273-fig3-data1-v1.xlsx
-
Figure 3—source data 2
Cell morphology (cell length and width) data for Figure 3D.
- https://cdn.elifesciences.org/articles/99273/elife-99273-fig3-data2-v1.xlsx
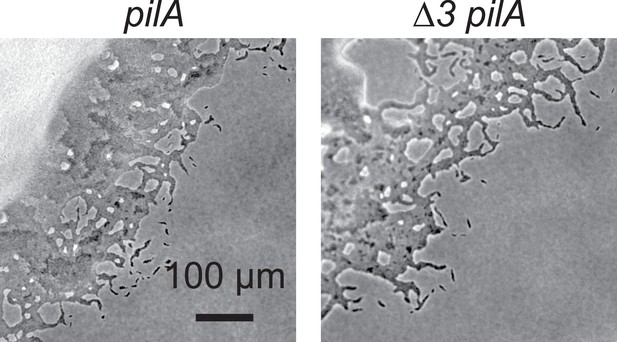
Moderate changes in cell dimensions do not affect gliding motility significantly.
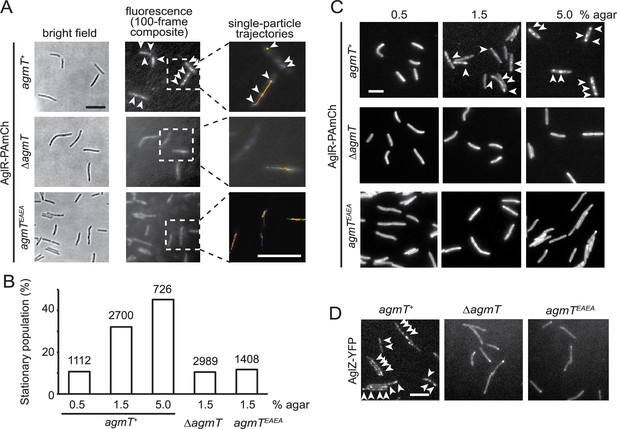
AgmT and its lytic transglycosylase (LTG) activity are essential for proper bacterial focal adhesion complex (bFAC) assembly.
(A) Overall distribution of AglR-PAmCherry particles on 1.5% agar surface is displayed using the composite of 100 consecutive frames taken at 100-ms intervals. Single-particle trajectories of AglR-PAmCherry (AglR-PAmCh) were generated from the same frames. Individual trajectories are distinguished by colors. Deleting agmT or disabling the transglycosylase activity of AgmT (AgmTEAEA) decrease the stationary population of AglR particles. (B) The stationary population of AglR-PAmCherry particles, which reflects the AglR molecules in bFACs, changes in response to substrate hardness (controlled by agar concentration) and the presence and function of AgmT. The total number of particles analyzed is shown on top of each plot. (C) AglR fails to assemble into bFACs in the absence of active AgmT, even on 5.0% agar surfaces. (D) AgmT and its LTG activity also support the assembly of AglZ into bFACs. White arrows point to bFACs. Scale bars, 5 µm.

AgmT does not assemble into bacterial focal adhesion complexes (bFACs) but connects bFACs to peptidoglycan (PG).
(A) Immunoblotting using M. xanthus cell lysates and an anti-mCherry antibody shows that PAmCherry (PAmCh)-labeled AgmT and AgmTEAEA (592 amino acids) accumulate as full-length proteins. AgmT is significantly more abundant than the PAmCherry-labeled motor protein AglR (498 amino acids). The bacterial actin homolog MreB visualized using an MreB antibody is shown as a loading control. (B) AgmT does not aggregate into bFACs. (C) The lytic transglycosylase (LTG) activity of AgmT is required for connecting bFACs to PG and expressing the E. coli LTG MltG (MltGEc) restores PG binding by bFACs in cells that lack AgmT but not in the ones that express an inactive AgmT variant (AgmTEAEA). AglR-PAmCherry was detected using an anti-mCherry antibody to mark the presence of bFACs that co-precipitate with PG-containing (lysozyme−) pellets. Lysates from the cells that express AglR-PAmCherry in different genetic backgrounds were pelleted by centrifugation in the presence and absence of lysozyme. The loading control of AglR-PAmCherry in the whole cell is shown as ‘total’.
-
Figure 5—source data 1
The original file of the full, raw, and unedited blot for the upper panel of Figure 5A.
- https://cdn.elifesciences.org/articles/99273/elife-99273-fig5-data1-v1.pdf
-
Figure 5—source data 2
The original file of the full, raw, and unedited blot for the lower panel of Figure 5A.
- https://cdn.elifesciences.org/articles/99273/elife-99273-fig5-data2-v1.pdf
-
Figure 5—source data 3
The original file of the full, raw, and unedited blot for the upper panel of Figure 5C.
- https://cdn.elifesciences.org/articles/99273/elife-99273-fig5-data3-v1.pdf
-
Figure 5—source data 4
The original file of the full, raw, and unedited blot for the middle panel of Figure 5C.
- https://cdn.elifesciences.org/articles/99273/elife-99273-fig5-data4-v1.pdf
-
Figure 5—source data 5
The original file of the full, raw, and unedited blot for the lower panel of Figure 5C.
- https://cdn.elifesciences.org/articles/99273/elife-99273-fig5-data5-v1.pdf
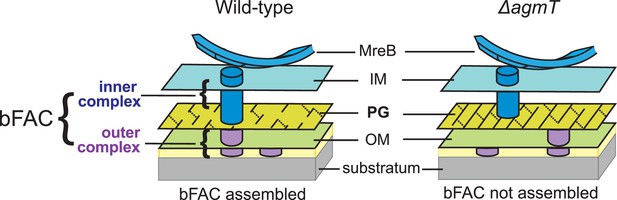
A possible mechanism by which AgmT connect bFACs to PG.
AgmT could generate short glycan strands through its lytic transglycosylase (LTG) activity and thus uniquely modify the overall structure of M. xanthus PG, such as producing small pores that retard and retain the inner subcomplexes of bFACs. Likewise, the M. xanthus mutants that lack active AgmT could produce PG with increased strand length, which precludes bFACs from binding to the cell wall.
Videos
Gliding motility of the ΔagmT pilA− cells.
Timelapse was captured at 10-s intervals and the video plays at 10 frames/s (100× speedup).
Gliding motility of the pilA− cells.
Timelapse was captured at 10-s intervals and the video plays at 10 frames/s (100× speedup).
Tables
| Reagent type (species) or resource | Designation | Source or reference | Identifiers | Additional information |
|---|---|---|---|---|
| Gene (M. xanthus) | agmT (K1515_04910) | Aramayo and Nan, 2022 | MXAN_RS31980 MXAN_6607 | MltG/YceG LTG family 5 |
| Gene (M. xanthus) | K1515_37860 | Aramayo and Nan, 2022 | MXAN_RS00570 MXAN_0114 | Slt/MltE LTG family 1 |
| Gene (M. xanthus) | K1515_37385 | Aramayo and Nan, 2022 | MXAN_RS01035 MXAN_0210 | Slt/MltE LTG family 1 |
| Gene (M. xanthus) | K1515_34725 | Aramayo and Nan, 2022 | MXAN_RS03640 MXAN_0754 | LTG family unknown |
| Gene (M. xanthus) | K1515_24775 | Aramayo and Nan, 2022 | MXAN_RS12365 MXAN_2558 | RlpA LTG family 6 |
| Gene (M. xanthus) | K1515_22185 | Aramayo and Nan, 2022 | MXAN_RS14935 MXAN_3081 | Slt/MltE LTG family 1 |
| Gene (M. xanthus) | K1515_20905 | Aramayo and Nan, 2022 | MXAN_RS16205 MXAN_3344 | Slt/MltE LTG family 1 |
| Gene (M. xanthus) | K1515_20820 | Aramayo and Nan, 2022 | MXAN_RS16290 MXAN_3363 | Slt/MltE LTG family 1 |
| Gene (M. xanthus) | K1515_17460 | Aramayo and Nan, 2022 | MXAN_RS19615 MXAN_4034 | Slt/MltE LTG family 1 |
| Gene (M. xanthus) | K1515_14545 | Aramayo and Nan, 2022 | MXAN_RS22465 MXAN_4628 | Slt/MltE LTG family 1 |
| Gene (M. xanthus) | K1515_09355 | Aramayo and Nan, 2022 | MXAN_RS27580 MXAN_5690 | Slt/MltD/MltE, LTG family 1 |
| Gene (M. xanthus) | K1515_06035 | Aramayo and Nan, 2022 | MXAN_RS30855 MXAN_6370 | Slt/MltE LTG family 1 |
| Gene (M. xanthus) | K1515_01490 | Aramayo and Nan, 2022 | MXAN_RS35365 MXAN_7308 | MltA LTG family 2 |
| Gene (M. xanthus) | K1515_01440 | Aramayo and Nan, 2022 | MXAN_RS35415 MXAN_7318 | Slt/MltD/MltE, LTG family 1 |
| Gene (E. coli) | mltG | UniProtKB | ec:4.2.2.29 | |
| Strain, strain background (M. xanthus) | DZ2 | Campos and Zusman, 1975 | DZ2 | Wild-type strain |
| Strain, strain background (M. xanthus) | aglR-PAmCherry | Nan et al., 2013 | N/A | |
| Strain, strain background (M. xanthus) | Δ3 (Δpbp1a1 Δpbp1a2 pbp1c::kan) | Zhang et al., 2023 | BN311 | |
| Strain, strain background (M. xanthus) | aglZ-yfp::kan | Mignot et al., 2007 | TM7 | |
| Strain, strain background (M. xanthus) | Δ3 (Δpbp1a1 Δpbp1a2 pbp1c::kan) pilA::tet | This paper | BN328 | Request from Nan lab |
| Strain, strain background (M. xanthus) | ΔagmT | This paper | BN329 | Request from Nan lab |
| Strain, strain background (M. xanthus) | ΔagmT pilA::tet | This paper | BN330 | Request from Nan lab |
| Strain, strain background (M. xanthus) | agmTEAEA | This paper | BN331 | Request from Nan lab |
| Strain, strain background (M. xanthus) | agmTEAEA pilA::tet | This paper | BN332 | Request from Nan lab |
| Strain, strain background (M. xanthus) | ΔaglR pilA::tet | This paper | BN333 | Request from Nan lab |
| Strain, strain background (M. xanthus) | agmT-PAmCherry | This paper | BN334 | Request from Nan lab |
| Strain, strain background (M. xanthus) | agmTEAEA-PAmCherry | This paper | BN335 | Request from Nan lab |
| Strain, strain background (M. xanthus) | agmT-PAmCherry pilA::tet | This paper | BN336 | Request from Nan lab |
| Strain, strain background (M. xanthus) | ΔagmT pilA::tet pMR3679 | This paper | BN337 | Request from Nan lab |
| Strain, strain background (M. xanthus) | ΔagmT pMR3679-mltGEC | This paper | BN338 | Request from Nan lab |
| Strain, strain background (M. xanthus) | ΔagmT pMR3679-mltGEC pilA::tet | This paper | BN339 | Request from Nan lab |
| Strain, strain background (M. xanthus) | agmTEAEA pMR3679-mltGEC | This paper | BN340 | Request from Nan lab |
| Strain, strain background (M. xanthus) | agmTEAEA pMR3679-mltGEC pilA::tet | This paper | BN341 | Request from Nan lab |
| Strain, strain background (M. xanthus) | aglR-PAmCherry ΔagmT | This paper | BN342 | Request from Nan lab |
| Strain, strain background (M. xanthus) | aglR-PAmCherry agmTEAEA | This paper | BN343 | Request from Nan lab |
| Strain, strain background (M. xanthus) | aglZ-yfp::kan ΔagmT | This paper | BN344 | Request from Nan lab |
| Strain, strain background (M. xanthus) | aglZ-yfp::kan agmTEAEA | This paper | BN345 | Request from Nan lab |
| Strain, strain background (M. xanthus) | aglR-PAmCherry aglZ-yfp::kan | This paper | BN346 | Request from Nan lab |
| Strain, strain background (M. xanthus) | K1515_37860::kan pilA::tet | This paper | BN347 | Request from Nan lab |
| Strain, strain background (M. xanthus) | K1515_37385::kan pilA::tet | This paper | BN348 | Request from Nan lab |
| Strain, strain background (M. xanthus) | K1515_34725::kan pilA::tet | This paper | BN349 | Request from Nan lab |
| Strain, strain background (M. xanthus) | ΔK1515_24775 pilA::tet | This paper | BN350 | Request from Nan lab |
| Strain, strain background (M. xanthus) | K1515_22185::kan pilA::tet | This paper | BN351 | Request from Nan lab |
| Strain, strain background (M. xanthus) | K1515_20905::kan pilA::tet | This paper | BN352 | Request from Nan lab |
| Strain, strain background (M. xanthus) | K1515_20820::kan pilA::tet | This paper | BN353 | Request from Nan lab |
| Strain, strain background (M. xanthus) | K1515_17460::kan pilA::tet | This paper | BN354 | Request from Nan lab |
| Strain, strain background (M. xanthus) | K1515_14545::kan pilA::tet | This paper | BN355 | Request from Nan lab |
| Strain, strain background (M. xanthus) | K1515_09355::kan pilA::tet | This paper | BN356 | Request from Nan lab |
| Strain, strain background (M. xanthus) | K1515_06035::kan pilA::tet | This paper | BN357 | Request from Nan lab |
| Strain, strain background (M. xanthus) | K1515_01490::kan pilA::tet | This paper | BN358 | Request from Nan lab |
| Strain, strain background (M. xanthus) | K1515_01440::kan pilA::tet | This paper | BN359 | Request from Nan lab |
| Antibody (Anti-mCherry) | Rabbit polyclonal | Rockland Immunochemicals | 600-401-P16 | (1:2000) |
| Antibody (Anti-MreB) | Rabbit polyclonal | Mauriello et al., 2010 | (1:20,000) | |
| Antibody (anti-rabbit IgG H+L (HRP)) | Goat polyclonal | Fisher Scientific | 31460 | (1:20,000) |
| Software, algorithm | MATLAB | MathWorks | ||
| Software, algorithm | Cell morphology analysis algorithm | Zhang et al., 2020; Zhang et al., 2023 | DOI: 10.5281/zenodo.8234126 | |
| Software, algorithm | Single-particle analysis algorithm | Fu et al., 2018; Zhang et al., 2023 | DOI: 10.5281/zenodo.8234126 | |
| Software, algorithm | TrackMate 7 | Ershov et al., 2022 | ||
| Software, algorithm | ImageJ | https://imagej.net |





