Executioner caspase is proximal to Fasciclin 3 which facilitates non-lethal activation in Drosophila olfactory receptor neurons
Figures
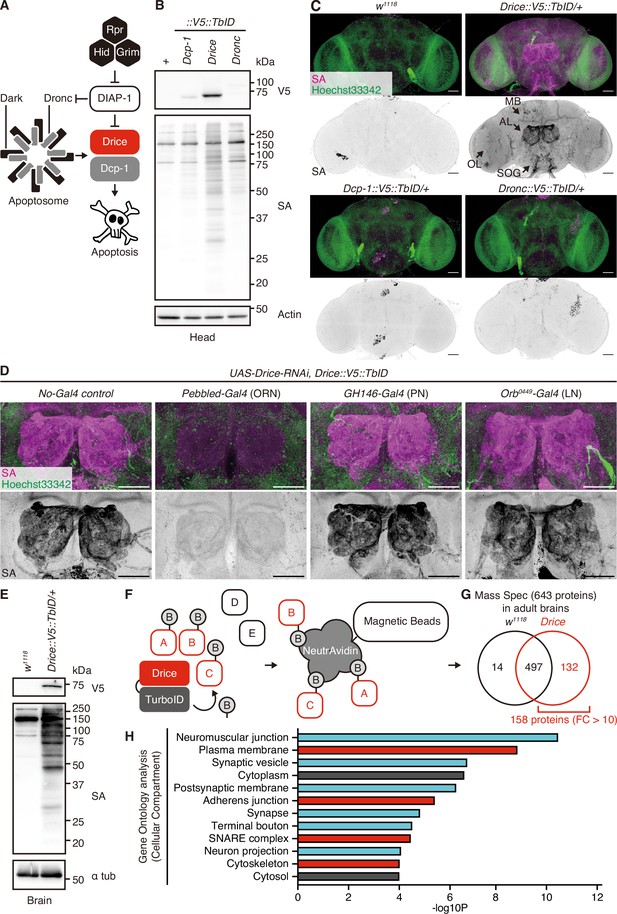
Expression patterns and proximal proteins of Drice in the adult brain.
(A) A schematic diagram of Drosophila apoptosis signaling. (B) Western blot of expression of C-terminally V5::TurboID knocked-in tagged caspases in adult male heads. Biotinylated proteins are detected by streptavidin (SA). (C) Representative images of adult male brains. Expression patterns of each C-terminally V5::TurboID knocked-in tagged caspase are visualized using SA (magenta). Nuclei are visualized by Hoechst 33342 (green). Arrows indicate the mushroom body (MB), antennal lobe (AL), optic lobe (OL), and subesophageal ganglion (SOG), respectively. Scale bar: 50 µm. (D) Representative images of ALs in the adult male brains. Expression patterns of Drice are visualized using SA (magenta). Nuclei are visualized by Hoechst 33342 (green). Scale bar: 50 µm. (E) Western blot of biotinylated proteins labeled by Drice::V5::TurboID extracted from adult male brains. Biotinylated proteins are detected by SA. (F) A schematic diagram of the TurboID-mediated identification of proximal proteins. Drice-proximal proteins are promiscuously labeled in vivo with the administration of biotin. Then, biotinylated proteins are purified using NeutrAvidin magnetic beads and are subsequently analyzed by mass spectrometry. (G) A summary of mass spectrometry analysis of proteins of the adult male brains. Among 643 proteins identified, 158 proteins were detected as highly specific proteins to Drice::V5::TurboID flies compared to wild-type flies (FC [Drice::V5::TurboID/w1118]>10). (H) Gene Ontology analysis [Cellular Compartment] of 158 Drice-proximal proteins (FC [Drice::V5::TurboID/w1118]>10). Cyan: neuronal-related fraction; red: membrane fraction.
-
Figure 1—source data 1
Uncropped raw blot images of Figure 1B and E.
- https://cdn.elifesciences.org/articles/99650/elife-99650-fig1-data1-v2.zip
-
Figure 1—source data 2
Uncropped annotated blot images of Figure 1B and E.
- https://cdn.elifesciences.org/articles/99650/elife-99650-fig1-data2-v2.zip

Expression patterns of Drice in the adult brain in males and females.
(A) Representative images of adult male and female brains. Expression patterns of C-terminally V5::TurboID knocked-in tagged Drice are visualized using streptavidin (SA, magenta). Nuclei are visualized by Hoechst 33342 (green). Scale bar: 50 µm. (B) Representative images of antennal lobes in the adult female brains. Expression patterns of Drice are visualized using SA (magenta). Nuclei are visualized by Hoechst 33342 (green). Scale bar: 50 µm.
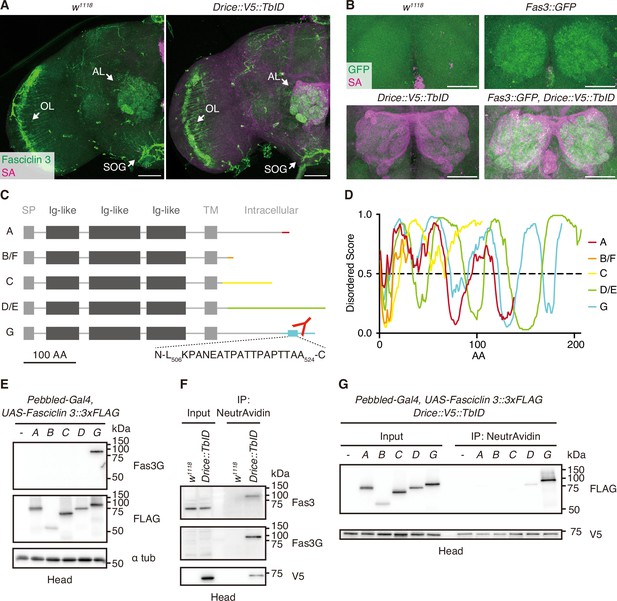
A specific isoform of Fasciclin 3 is in proximity to Drice.
(A) Representative expression patterns of C-terminally V5::TurboID knocked-in tagged Drice (streptavidin [SA]; magenta) and Fasciclin 3 (Fas3, anti-Fasciclin 3 antibody staining; green) in the adult male brains. Arrows indicate the antennal lobe (AL), optic lobe (OL), and subesophageal ganglion (SOG), respectively. Scale bar: 50 µm. (B) Representative expression patterns of C-terminally V5::TurboID knocked-in tagged Drice (SA; magenta) and Fas3 (anti-GFP antibody staining; green) in the ALs of the adult male brains. Scale bar: 50 µm. (C) Schematic protein structures of Fas3 isoforms. Intracellular regions differ from each other. A peptide region used to raise anti-Fas3G antibody is shown. Signal peptides (SPs), immunoglobulin-like (Ig-like) domains, and transmembrane (TM) domains are shown in boxes. (D) IDR predictions of intracellular regions of each protein isoform. Regions wherein the score is more than 0.5 are predicted to be disordered. (E) Western blot of proteins extracted from adult heads overexpressing each 3xFLAG-tagged Fas3 isoform. Anti-Fas3G antibody specifically detects Fas3 isoform G. (F) Western blot of endogenous Fas3 expressed in adult male heads. Drice-proximal proteins biotinylated by C-terminally knocked-in tagged V5::TurboID are purified using NeutrAvidin. (G) Western blot of 3xFLAG-tagged Fas3 isoforms overexpressed in adult head. Drice-proximal proteins biotinylated by C-terminally knocked-in tagged V5::TurboID are purified using NeutrAvidin.
-
Figure 2—source data 1
Uncropped raw blot images of Figure 2E, F, and G.
- https://cdn.elifesciences.org/articles/99650/elife-99650-fig2-data1-v2.zip
-
Figure 2—source data 2
Uncropped annotated blot images of Figure 2E, F, and G.
- https://cdn.elifesciences.org/articles/99650/elife-99650-fig2-data2-v2.zip

Predicted protein complexes of Drice and each Fas3 isoform.
(A) Predicted aligned error (PAE) plots of protein complex of Drice and each Fasciclin 3 (Fas3) isoform generated by AlphaFold2-Multimer. In the Y axis, A represents Drice and B represents each Fas3 isoform. Numbers in the X axis represent amino acid position. The intracellular region of each Fas3 isoform is represented by the orange bar. Potential interaction sites predicted by low PAE scores in Drice-Fas3A/G are highlighted by blue rectangles.

Biochemical property of Fas3s with caspases.
(A) Co-immunoprecipitation of 3xFLAG-tagged Fasciclin 3s (Fas3s) with V5-tagged Drice extracted from adult male heads. 3xFLAG-tagged Fas3s are immunoprecipitated using an anti-FLAG antibody. Biotinylated proteins labeled by Drice::V5::TurboID are detected by streptavidin (SA). (B) Western blot for 3xFLAG-tagged Fas3s expressed in Drosophila S2 cells. Apoptosis was induced by treating Drosophila S2 cells with cycloheximide (CHX). Induction of caspase activation is confirmed by the cleavage of endogenous Lamin B.
-
Figure 2—figure supplement 2—source data 1
Uncropped raw blot images of Figure 2—figure supplement 2A and B.
- https://cdn.elifesciences.org/articles/99650/elife-99650-fig2-figsupp2-data1-v2.zip
-
Figure 2—figure supplement 2—source data 2
Uncropped annotated blot images of Figure 2—figure supplement 2A and B.
- https://cdn.elifesciences.org/articles/99650/elife-99650-fig2-figsupp2-data2-v2.zip
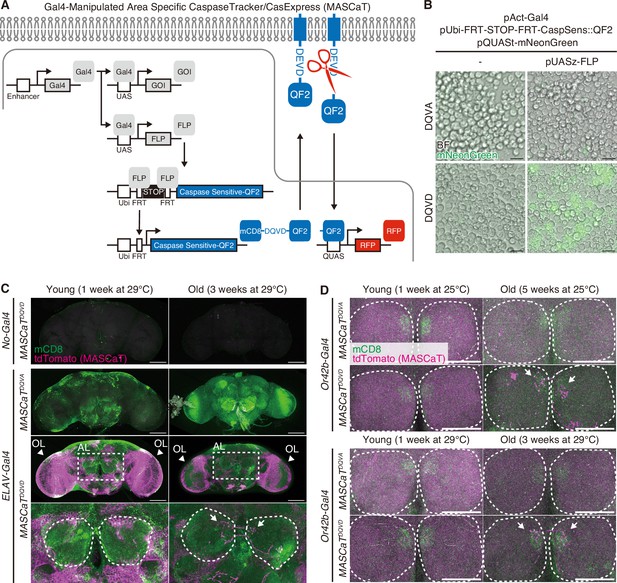
A Gal4-Manipulated Area Specific CaspaseTracker/CasExpress (MASCaT).
(A) A schematic diagram of MASCaT expressed using the Gal4/UAS system with FLP-mediated recombination. Caspase-sensitive-QF2 is activated by caspase-mediated cleavage at the plasma membrane. Cleaved QF2 translocates to the nucleus to induce reporter protein expression downstream of QUAS sequence. (B) Representative images of Drosophila S2 cells expressing caspase-sensitive QF2 probes by the Gal4/UAS system with FLP-mediated recombination. Scale bar: 20 µm. (C) Representative images of adult male brains of young (1 week of age) and old (3 weeks of age) flies raised at 29°C expressing MASCaT probe (mCD8; green). Caspase activation is visualized by MASCaT-induced tdTomato (magenta) expression. Arrowheads indicate optic lobes (OLs). Magnified images (white rectangle) show antennal lobes (ALs; white circles). Arrows indicate age-dependent caspase activation at the ALs. Scale bar: 100 µm. (D) Representative images of adult male brains of young (1 week of age raised at 25°C or 29°C) and old (5 weeks of age raised at 25°C or 3 weeks of age raised at 29°C) expressing MASCaT probe (mCD8; green). Caspase activation is visualized by MASCaT-induced tdTomato (magenta) expression. White circles show ALs. Arrows indicate age-dependent caspase activation at the ALs. Scale bar: 50 µm.

Overexpression of mCD8::DQVD::QF2 probes in Drosophila S2 cells.
(A) A schematic diagram of Gal4-Manipulated Area Specific CaspaseTracker/CasExpress (MASCaT) expressed directly using the Gal4/UAS system. Caspase-sensitive QF2 is activated by caspase-mediated cleavage at the plasma membrane. In theory, only cleaved QF2 translocates to the nucleus to induce reporter protein expression downstream of QUAS sequence. (B) Representative images of Drosophila S2 cells expressing caspase-sensitive QF2 probes directly by the Gal4/UAS system. A pan-caspase inhibitor, zVAD-fmk, is treated to inhibit caspase activity. Scale bar: 20 µm.
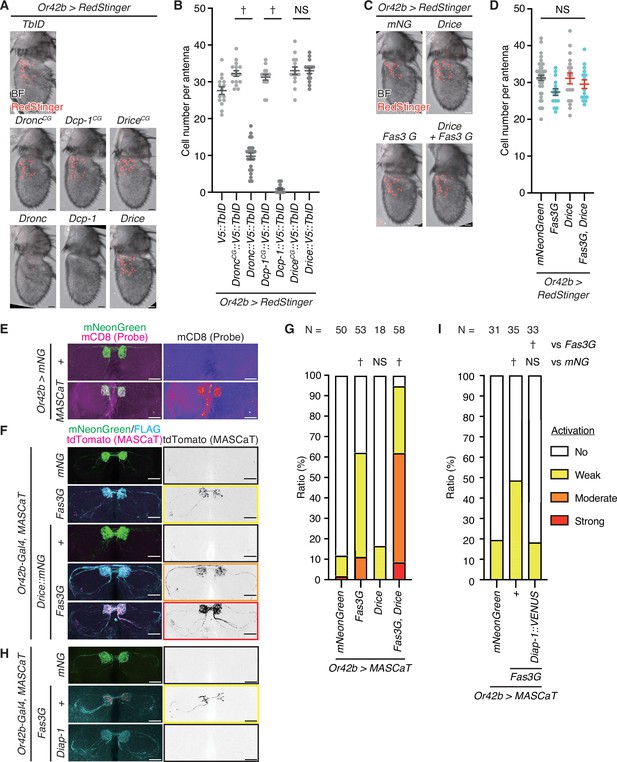
Fasciclin 3 isoform G overexpression enhances non-lethal caspase activation.
(A) Representative images of the adult male antennae of 1-week-old flies raised at 29°C. Cell bodies are visualized using RedStinger (red). Scale bar: 20 µm. (B) Quantification of cell number of (A). Data are presented as mean ± SEM. p-Values were calculated using one-way analysis of variance (ANOVA) with Bonferroni’s correction for selected pairs. NS: p>0.05, †: p<0.05. Sample sizes: V5::TbID (N=15), DroncCG:::V5::TbID (N=16), Dronc:::V5::TbID (N=23), Dcp-1CG::V5::TbID (N=15), Dcp-1::V5::TbID (N=16), DriceCG::V5::TbID (N=15), Drice::V5::TbID (N=15). (C) Representative images of the adult male antennae of 1-week-old flies raised at 29°C. Cell bodies are visualized using RedStinger (red). Scale bar: 20 µm. (D) Quantification of cell number of (C). Data are presented as mean ± SEM. Light blue dots represent Fas3G overexpression conditions while red bars represent Drice overexpression conditions. p-Values were calculated using one-way ANOVA with Bonferroni’s correction for every pair. NS: p>0.05. Sample sizes: mNeonGreen (N=44), Fasciclin 3 isoform G (Fas3G; N=15), Drice (N=18), Fas3G+Drice (N=15). (E) Representative images of the antennal lobes (ALs) of male brains of 1-week-old flies raised at 29°C expressing MASCaT probe (mCD8; magenta). Expression of the probes is restricted to the Or42b-Gal4 positive (mNeonGreen; green) regions. Scale bar: 30 µm. (F) Representative images of the ALs of brains of 1-week-old flies raised at 29°C with caspase activation visualized by MASCaT-induced tdTomato (magenta) expression. Expression of mNeonGreen (green) or 3xFLAG-tagged Fas3G (cyan) is simultaneously visualized. Scale bar: 30 µm. (G) Quantifications of caspase activity detected by MASCaT of (F). p-Values were calculated using chi-square test with Bonferroni’s correction using mNeonGreen as control. NS: p>0.05, †: p<0.05. Sample sizes are shown in the graph. (H) Representative images of the ALs of brains of 1-week-old flies raised at 29°C with caspase activation visualized by MASCaT-induced tdTomato (magenta) expression. Expression of mNeonGreen (green) or 3xFLAG-tagged Fas3G (cyan) is simultaneously visualized. Scale bar: 30 µm. (I) Quantifications of caspase activity detected by MASCaT of (H). p-Values were calculated using chi-square test with Bonferroni’s correction using mNeonGreen and Fas3G as controls, respectively. NS: p>0.05, †: p<0.05. Sample sizes are shown in the graph.
-
Figure 4—source data 1
Data used for graphs presented in Figure 4B, D, G, and I.
- https://cdn.elifesciences.org/articles/99650/elife-99650-fig4-data1-v2.xlsx

Loss-of-function analysis of Fas3G.
(A) Gene structure of Fasciclin 3 (Fas3). CDS (orange) and UTR (gray) are shown in boxes. Target sequence of Fas3G-shRNA is shown in red. (B) Western blot for 3xFLAG-tagged Fas3s expressed in Drosophila S2 cells with Fas3G-shRNA. (C) Representative images of the antennal lobes (ALs) of brains of 1-week-old flies raised at 29°C with caspase activation visualized by Gal4-Manipulated Area Specific CaspaseTracker/CasExpress (MASCaT) probe (mCD8; green)-induced tdTomato (magenta) expression. Scale bar: 30 µm. (D) Quantifications of caspase activity detected by MASCaT of (C). p-Values were calculated using chi-square test with Bonferroni’s correction using LacZ-RNAi as control. NS: p>0.05. Sample sizes are shown in the graph. (E) Representative images of the ALs of brains of 1-week-old flies expressing mNeonGreen-tagged Drice (green) and 3xFLAG-tagged Fas3G (magenta). Scale bar: 50 µm.
-
Figure 4—figure supplement 1—source data 1
Uncropped raw blot images of Figure 4—figure supplement 1B.
- https://cdn.elifesciences.org/articles/99650/elife-99650-fig4-figsupp1-data1-v2.zip
-
Figure 4—figure supplement 1—source data 2
Uncropped annotated blot images of Figure 4—figure supplement 1B.
- https://cdn.elifesciences.org/articles/99650/elife-99650-fig4-figsupp1-data2-v2.zip
-
Figure 4—figure supplement 1—source data 3
Data used for graphs presented in Figure 4—figure supplement 1D.
- https://cdn.elifesciences.org/articles/99650/elife-99650-fig4-figsupp1-data3-v2.xlsx
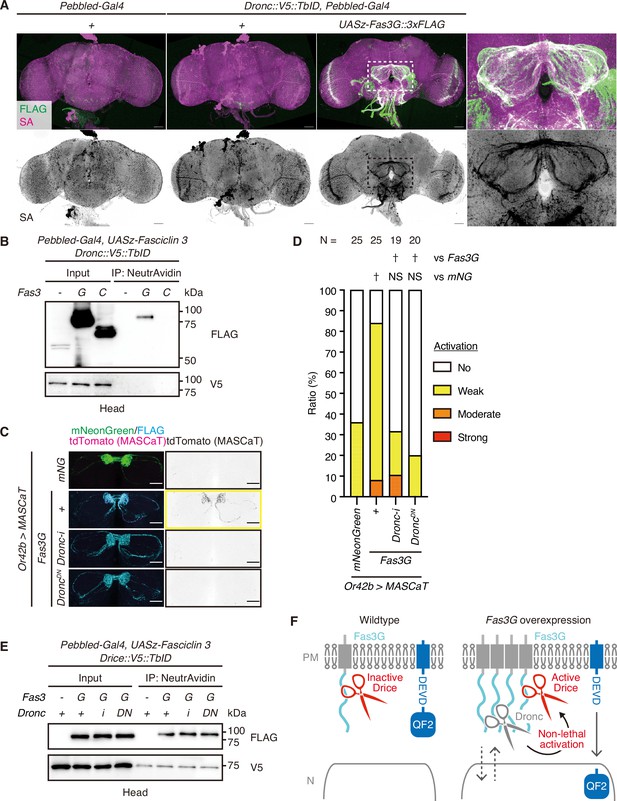
Fasciclin 3 isoform G overexpression enhances non-lethal caspase activation in a Dronc-dependent manner.
(A) Representative images of adult male brains. Expression patterns of C-terminally V5::TurboID knocked-in tagged Dronc are visualized using streptavidin (SA; magenta) with or without the expression of 3xFLAG-tagged Fasciclin isoform G (Fas3G; green). Magnified images (white rectangle) show antennal lobes (ALs). Scale bar: 50 µm. (B) Western blot of adult male heads. Dronc-proximal proteins biotinylated by C-terminally knocked-in tagged V5::TurboID are purified using NeutrAvidin. (C) Representative images of the AL of brains of 1-week-old flies raised at 29°C with caspase activation visualized by Gal4-Manipulated Area Specific CaspaseTracker/CasExpress (MASCaT)-induced tdTomato (magenta) expression. Expression of mNeonGreen (green) or 3xFLAG-tagged Fas3G (cyan) is simultaneously visualized. Scale bar: 30 µm. (D) Quantifications of caspase activity detected using MASCaT of (C). p-Values were calculated using chi-square test with Bonferroni’s correction using mNeonGreen and Fas3G as controls, respectively. NS: p>0.05, †: p<0.05. Sample sizes are shown in the graph. (E) Western blot of adult male heads. Drice-proximal proteins biotinylated by C-terminally knocked-in tagged V5::TurboID are purified using NeutrAvidin. (F) A schematic model of subcellularly restricted non-lethal caspase activation at the plasma membrane (PM). Inactive Drice is in proximity to Fas3G in wild-type condition. Fas3G overexpression induces Dronc expression. Then, Dronc comes close to Fas3G and activates Drice. Activated Drice non-lethally cleaves substrate near PM, including MASCaT probes.
-
Figure 5—source data 1
Uncropped raw blot images of Figure 5B and E.
- https://cdn.elifesciences.org/articles/99650/elife-99650-fig5-data1-v2.zip
-
Figure 5—source data 2
Uncropped annotated blot images of Figure 5B and E.
- https://cdn.elifesciences.org/articles/99650/elife-99650-fig5-data2-v2.zip
-
Figure 5—source data 3
Data used for graphs presented in Figure 5D.
- https://cdn.elifesciences.org/articles/99650/elife-99650-fig5-data3-v2.xlsx
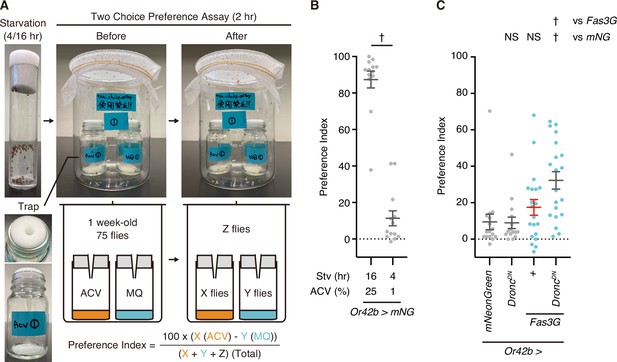
Fasciclin 3 isoform G overexpression-facilitated caspase activation regulates attraction behavior.
(A) Images and a schematic diagram of two-choice preference assay. Flies are starved for 4 or 16 hr before the assay. One-week-old young flies are left for 2 hr in the bottle with two traps. After the assay, preference index is calculated by counting the number of flies left in the trap with apple cider vinegar (ACV) as X, in the trap with MilliQ (MQ) water as Y, and other (field) compartments as Z. (B) Preference index of 1-week-old male flies raised at 29°C. Flies are starved for 16 or 4 hr and are subsequently tested in response to 25% or 1% ACV in a two-choice preference assay. Data are presented as mean ± SEM. p-Values were calculated using unpaired t-test. †: p<0.05. Sample sizes: 16 hr Stv/25% AC (N=13 [915 flies]), 4 hr Stv/1% ACV (N=13 [945 flies]). (C) Preference index of 1-week-old male flies raised at 29°C. Flies are starved for 4 hr and are subsequently tested in response to 1% ACV in a two-choice preference assay. Data are presented as mean ± SEM. Light blue dots represent Fas3G overexpression conditions, while a red bar indicates caspase-activated condition. p-Values were calculated using one-way analysis of variance (ANOVA) followed by Dunnets’ multiple comparison test using mNeonGreen and Fas3G as controls. NS: p>0.05, †: p<0.05. Sample sizes: mNeonGreen (N=16 [1159 flies]), DroncDN (N=15 [1099 flies]), Fas3G (N=20 [1419 flies]), Fas3G+DroncDN (N=20 [1318 flies]).
-
Figure 6—source data 1
Data used for graphs presented in Figure 6B and C.
- https://cdn.elifesciences.org/articles/99650/elife-99650-fig6-data1-v2.xlsx
Tables
| Reagent type (species) or resource | Designation | Source or reference | Identifiers | Additional information |
|---|---|---|---|---|
| Gene (Drosophila melanogaster) | Fas3-PA | FlyBase | FLYB: FBpp0080604 | |
| Gene (Drosophila melanogaster) | Fas3-PB | FlyBase | FLYB: FBpp0080605 | |
| Gene (Drosophila melanogaster) | Fas3-PC | FlyBase | FLYB: FBpp0111716 | |
| Gene (Drosophila melanogaster) | Fas3-PD | FlyBase | FLYB: FBpp0111717 | |
| Gene (Drosophila melanogaster) | Fas3-PE | FlyBase | FLYB: FBpp0290605 | |
| Gene (Drosophila melanogaster) | Fas3-PF | FlyBase | FLYB: FBpp0309467 | |
| Gene (Drosophila melanogaster) | Fas3-PG | FlyBase | FLYB: FBpp0311111 | |
| Gene (Drosophila melanogaster) | Drice | FlyBase | FLYB: FBpp0084848 | |
| Genetic reagent (Drosophila melanogaster) | w1118 | Shinoda et al., 2023 | Maintained in Masayuki Miura lab | |
| Genetic reagent (Drosophila melanogaster) | Drice::V5::TurboID | Shinoda et al., 2019 | FLYB: FBal0356244 | |
| Genetic reagent (Drosophila melanogaster) | Dcp-1::V5::TurboID | Shinoda et al., 2019 | FLYB: FBal0356245 | |
| Genetic reagent (Drosophila melanogaster) | Dronc::V5::TurboID | Shinoda et al., 2019 | FLYB: FBal0356243 | |
| Genetic reagent (Drosophila melanogaster) | Pebbled-Gal4 | Bloomington Drosophila Stock Center | RRID:BDSC_80570 | |
| Genetic reagent (Drosophila melanogaster) | Orb0449-Gal4 | Bloomington Drosophila Stock Center | RRID:BDSC_63325 | |
| Genetic reagent (Drosophila melanogaster) | GH146-Gal4 | Bloomington Drosophila Stock Center | RRID:BDSC_30026 | |
| Genetic reagent (Drosophila melanogaster) | UAS-Drice-RNAi | Vienna Drosophila Resource Center | FLYB: FBst0457273 | |
| Genetic reagent (Drosophila melanogaster) | Fas3::EGFP::FlAsH:: StrepII::TEV::3xFLAG | Bloomington Drosophila Stock Center | RRID:BDSC_59809 | |
| Genetic reagent (Drosophila melanogaster) | UASz-Fas3A::3xFLAG | This paper | Integrated into attP2 landing site | |
| Genetic reagent (Drosophila melanogaster) | UASz-Fas3B::3xFLAG | This paper | Integrated into attP2 landing site | |
| Genetic reagent (Drosophila melanogaster) | UASz-Fas3C::3xFLAG | This paper | Integrated into attP2 landing site | |
| Genetic reagent (Drosophila melanogaster) | UASz-Fas3D::3xFLAG | This paper | Integrated into attP2 landing site | |
| Genetic reagent (Drosophila melanogaster) | UASz-Fas3G::3xFLAG | This paper | Integrated into attP2 landing site | |
| Genetic reagent (Drosophila melanogaster) | Ubi-FRT-STOP-FRT- mCD8::DQVD::QF2 | This paper | Integrated into attP40 landing site | |
| Genetic reagent (Drosophila melanogaster) | Ubi-FRT-STOP-FRT- mCD8::DQVA::QF2 | This paper | Integrated into attP40 landing site | |
| Genetic reagent (Drosophila melanogaster) | UAS-FLP | Kyoto Stock Center | DGRC Number: 107788 | |
| Genetic reagent (Drosophila melanogaster) | QUAS-tdTomato::3xHA | Bloomington Drosophila Stock Center | RRID:BDSC_30005 | |
| Genetic reagent (Drosophila melanogaster) | ELAV-Gal4 | Bloomington Drosophila Stock Center | RRID:BDSC_8765 | |
| Genetic reagent (Drosophila melanogaster) | Or42b-Gal4 | Bloomington Drosophila Stock Center | RRID:BDSC_9971 | |
| Genetic reagent (Drosophila melanogaster) | UAS-RedStinger | Bloomington Drosophila Stock Center | RRID:BDSC_8547 | |
| Genetic reagent (Drosophila melanogaster) | UASz-3xFLAG:: mNeonGreen | This paper | Integrated into attP40 landing site | |
| Genetic reagent (Drosophila melanogaster) | UASz-Drice::myc:: mNeonGreen | This paper | Integrated into attP40 landing site | |
| Genetic reagent (Drosophila melanogaster) | UASz-V5::TurboID | This paper | Integrated into attP2 landing site | |
| Genetic reagent (Drosophila melanogaster) | UASz-Dronc::V5::TurboID | This paper | Integrated into attP2 landing site | |
| Genetic reagent (Drosophila melanogaster) | UASz-DroncCG::V5::TurboID | This paper | Integrated into attP2 landing site | |
| Genetic reagent (Drosophila melanogaster) | UASz-Dcp-1::V5::TurboID | This paper | Integrated into attP2 landing site | |
| Genetic reagent (Drosophila melanogaster) | UASz-Dcp-1CG::V5::TurboID | This paper | Integrated into attP2 landing site | |
| Genetic reagent (Drosophila melanogaster) | UASz-Drice::V5::TurboID | This paper | Integrated into attP2 landing site | |
| Genetic reagent (Drosophila melanogaster) | UASz-DriceCG::V5::TurboID | This paper | Integrated into attP2 landing site | |
| Genetic reagent (Drosophila melanogaster) | UAS-Diap-1::VENUS | Koto et al., 2009 | FLYB: FBal0244058 | |
| Genetic reagent (Drosophila melanogaster) | UAS-Dronc-RNAi | Kanuka et al., 2005 | FLYB: FBal0191075 | |
| Genetic reagent (Drosophila melanogaster) | UAS-DroncDN::EGFP | Igaki, 2002 | FLYB: FBal0104748 | |
| Genetic reagent (Drosophila melanogaster) | UAS-LacZ-RNAi | Kennerdell and Carthew, 2000 | FLYB: FBal0196940 | Gift from Dr. Richard Carthew |
| Genetic reagent (Drosophila melanogaster) | UAS-Fas3-RNAi#1 | Vienna Drosophila Resource Center | FLYB: FBst0471491 | |
| Genetic reagent (Drosophila melanogaster) | UAS-Fas3-RNAi#2 | Vienna Drosophila Resource Center | FLYB: FBst0458720 | |
| Genetic reagent (Drosophila melanogaster) | UAS-Fas3G-RNAi | This paper | Integrated into attP40 landing site | |
| Cell line (Drosophila melanogaster) | S2 | RIKEN BRC | Cat#: RCB1153; RRID:CVCL_Z232 | Maintained in Masayuki Miura lab |
| Antibody | anti-V5 (mouse monoclonal) | Invitrogen | Cat#: 46-0705; RRID:AB_2556564 | WB (1:5,000) |
| Antibody | anti-FLAG M2 (mouse monoclonal) | Sigma | Cat#: F1804; RRID:AB_262044 | WB (1:5,000), IHC (1:500) |
| Antibody | anti-alpha tubulin (DM1A) (mouse monoclonal) | Sigma | Cat#: T9026; RRID:AB_477593 | WB (1:10,000) |
| Antibody | anti-Actin (mouse monoclonal) | Sigma | Cat#: A4700; RRID:AB_476730 | WB (1:5,000) |
| Antibody | anti-Fasciclin 3 (7G10) (mouse monoclonal) | DSHB | Cat#: 7G10 anti-Fasciclin III; RRID:AB_528238 | WB (1:50), IHC (1:20) |
| Antibody | anti-Fasciclin3 isoform G (rabbit polyclonal) | This paper | WB (1:50) | |
| Antibody | anti-Drosophila Lamin B (mouse monoclonal) | DSHB | Cat#: ADL67.10; RRID:AB_528336 | WB (1:1,000) |
| Antibody | anti-GFP (GF090R) (rat monoclonal) | nacalai tesque | Cat#: 04404-26; RRID:AB_10013361 | IHC (1:100) |
| Antibody | anti-mCD8 (rat monoclonal) | Invitrogen | Cat#: MCD0800; RRID:AB_10392843 | IHC (1:50) |
| Antibody | anti-rabbit IgG HRP- conjugated (goat polyclonal) | CST | Cat#: 7074S; RRID:AB_2099233 | WB (1:5,000) |
| Antibody | anti-mouse IgG HRP- conjugated (goat polyclonal) | Promega | Cat#: W402B; RRID:AB_430834 | WB (1:5,000) |
| Antibody | anti-mouse IgG Alexa Fluor 488-conjugated (donkey polyclonal) | ThermoFisher Scientific | Cat#: A-21202; RRID:AB_141607 | IHC (1:500) |
| Antibody | anti-rat IgG Alexa Fluor 488-conjugated (donkey polyclonal) | ThermoFisher Scientific | Cat#: A-21208; RRID:AB_2535794 | IHC (1:500) |
| Antibody | anti-mouse IgG Alexa Fluor 647-conjugated (donkey polyclonal) | ThermoFisher Scientific | Cat#: A-31571; RRID:AB_162542 | IHC (1:500) |
| Antibody | anti-rat IgG Alexa Fluor 633-conjugated (goat polyclonal) | ThermoFisher Scientific | Cat#: A-21094; RRID:AB_2535749 | IHC (1:500) |
| Recombinant DNA reagent | pUASz1.0 (plasmid) | Drosophila Genomics Resource Center | Cat#: 1431; RRID:DGRC_1431 | |
| Recombinant DNA reagent | pUASz-Fasciclin 3 isoform A-3xFLAG (plasmid) | This paper | ||
| Recombinant DNA reagent | pUASz-Fasciclin 3 isoform B/F-3xFLAG (plasmid) | This paper | ||
| Recombinant DNA reagent | pUASz-Fasciclin 3 isoform C-3xFLAG (plasmid) | This paper | ||
| Recombinant DNA reagent | pUASz-Fasciclin 3 isoform D/E-3xFLAG (plasmid) | This paper | ||
| Recombinant DNA reagent | pUASz-Fasciclin 3 isoform G-3xFLAG (plasmid) | This paper | ||
| Recombinant DNA reagent | pAc5-STABLE2-neo (plasmid) | Addgene | Cat#: 32426; RRID:Addgene_32426 | |
| Recombinant DNA reagent | pAc5-Fasciclin 3 isoform A-3xFLAG (plasmid) | This paper | ||
| Recombinant DNA reagent | pAc5-Fasciclin 3 isoform B/F-3xFLAG (plasmid) | This paper | ||
| Recombinant DNA reagent | pAc5-Fasciclin 3 isoform C-3xFLAG (plasmid) | This paper | ||
| Recombinant DNA reagent | pAc5-Fasciclin 3 isoform D/E-3xFLAG (plasmid) | This paper | ||
| Recombinant DNA reagent | pAc5-Fasciclin 3 isoform G-3xFLAG (plasmid) | This paper | ||
| Recombinant DNA reagent | pAc5-V5-mNeonGreen (plasmid) | Shinoda et al., 2023 | ||
| Recombinant DNA reagent | pUASz-3xFLAG- mNeonGreen (plasmid) | This paper | ||
| Recombinant DNA reagent | pUASz-Drice-myc- mNeonGreen (plasmid) | This paper | ||
| Recombinant DNA reagent | V5-TurboID- NES_pcDNA3 (plasmid) | Addgene | Cat#: 107169; RRID:Addgene_107169 | |
| Recombinant DNA reagent | pUASz-V5- TurboID (plasmid) | This paper | ||
| Recombinant DNA reagent | pUASz-Dronc-V5- TurboID (plasmid) | This paper | ||
| Recombinant DNA reagent | pUASz-DroncC318G-V5- TurboID (plasmid) | This paper | ||
| Recombinant DNA reagent | pUASz-Dcp-1-V5- TurboID (plasmid) | This paper | ||
| Recombinant DNA reagent | pUASz-Dcp-1C196G-V5- TurboID (plasmid) | This paper | ||
| Recombinant DNA reagent | pUASz-Drice-V5- TurboID (plasmid) | This paper | ||
| Recombinant DNA reagent | pUASz-DriceC211G-V5- TurboID (plasmid) | This paper | ||
| Recombinant DNA reagent | pJFRC-28K (plasmid) | Gift from Dr. Hokuto Kazama | ||
| Recombinant DNA reagent | pBPQU (plasmid) | Kashio et al., 2016 | ||
| Recombinant DNA reagent | pJFRC-mCD8-DQVD-QF2 (plasmid) | This paper | ||
| Recombinant DNA reagent | pJFRC-mCD8-DQVA-QF2 (plasmid) | This paper | ||
| Recombinant DNA reagent | pUbi83 (plasmid) | Gift from Dr. Daiki Umetsu | ||
| Recombinant DNA reagent | pJFRC201-10xUAS-FRT- STOP-FRT-myr-smGFP-HA (plasmid) | Addgene | Cat#: 63166; RRID:Addgene_63166 | |
| Recombinant DNA reagent | pUbi-FRT-STOP-FRT- mCD8-DQVD-QF2 (plasmid) | This paper | ||
| Recombinant DNA reagent | pUbi-FRT-STOP-FRT- mCD8-DQVA-QF2 (plasmid) | This paper | ||
| Recombinant DNA reagent | pQUAST (plasmid) | Addgene | Cat#: 24349; RRID:Addgene_24349 | |
| Recombinant DNA reagent | pQUAST-mNeonGreen (plasmid) | This paper | ||
| Recombinant DNA reagent | pQUAST-FLPo (plasmid) | Addgene | Cat#: 24357; RRID:Addgene_24357 | |
| Recombinant DNA reagent | pUASz-FLPo (plasmid) | This paper | ||
| Recombinant DNA reagent | pWALIUM20 (plasmid) | Drosophila Genomics Resource Center | Cat#: 1472; RRID:DGRC_1472 | |
| Recombinant DNA reagent | pWALIUM20-Fas3G- shRNA (plasmid) | This paper | ||
| Peptide, recombinant protein | The peptide corresponding to the 506–524 aa (N-LKPANEATPATTPAPTTAA-C) of Fasciclin 3 isoform G | This paper | Generated by Eurofins Inc. | |
| Commercial assay or kit | Anti-FLAG M2 Magnetic Beads | Millipore | Cat#: M8823-1ML | |
| Commercial assay or kit | FG-NeutrAvidin beads | Tamagawa Seiki | Cat#: TAS8848 N1171 | |
| Commercial assay or kit | MycoStrip | InvivoGen | Cat#: rep-mys-10 | |
| Commercial assay or kit | Effectene Transfection Reagent | QIAGEN | Cat#: 301427 | |
| Chemical compound, drug | (+)-Biotin | WAKO | Cat#: 029-08713 | |
| Chemical compound, drug | Streptavidin HRP-conjugated | Invitrogen | Cat#: SA10001 | WB (1:10,000) |
| Chemical compound, drug | Hoechst 33342 | Invitrogen | Cat#: H3570 | IHC (8 µM) |
| Chemical compound, drug | Streptavidin-Cy2 | Jackson ImmunoResearch | Cat#: 016-220-084 | IHC (1:500) |
| Chemical compound, drug | Streptavidin-Cy5 | Jackson ImmunoResearch | Cat#: 016-170-084 | IHC (1:500) |
| Chemical compound, drug | SlowFade Gold antifade reagent | Invitrogen | Cat#: S36939 | |
| Chemical compound, drug | Cycloheximide | Sigma | Cat#: C7698 | |
| Chemical compound, drug | Apple cider vinegar | Mizkan | ||
| Software, algorithm | Fiji (ImageJ) | NIH Image | https://imagej.net/software/fiji/ | |
| Software, algorithm | Proteome Discoverer (PD) 2.2 | Thermo Fisher Scientific | ||
| Software, algorithm | DAVID | Huang et al., 2009 | https://davidbioinformatics.nih.gov/ | |
| Software, algorithm | PSIPRED | http://bioinf.cs.ucl.ac.uk/psipred/ | ||
| Software, algorithm | AlphaFold2-Multimer | Mirdita et al., 2022 | ColabFold v1.5.5: AlphaFold2 using MMseqs2 | |
| Software, algorithm | GraphPad Prism 8 | GraphPad |
Additional files
-
Supplementary file 1
Proximal protein list.
A list of the identified proximal proteins (Abundance ratio (Drice::V5::TurboID/Control) > 10). The UniProt Accessions, Gene Description, and Abundance Ratio (Drice::V5::TurboID/Control) are listed.
- https://cdn.elifesciences.org/articles/99650/elife-99650-supp1-v2.xlsx
-
Supplementary file 2
Gene Ontology (GO) analysis.
The result of GO analysis of Cellular Compartment for Drice-proximal proteins. GO term, p-values, Fold Enrichment, Count, and Genes are listed.
- https://cdn.elifesciences.org/articles/99650/elife-99650-supp2-v2.xlsx
-
Supplementary file 3
Fly stock list.
A list of the fly strains. Genotype, Map, Source or Reference, and Stock# are listed.
- https://cdn.elifesciences.org/articles/99650/elife-99650-supp3-v2.xlsx
-
Supplementary file 4
Detailed genotypes.
Detailed genotypes and their related figure # are presented.
- https://cdn.elifesciences.org/articles/99650/elife-99650-supp4-v2.xlsx
-
Supplementary file 5
Liquid chromatography (LC) settings.
Gradient settings for LC analysis are presented.
- https://cdn.elifesciences.org/articles/99650/elife-99650-supp5-v2.xlsx
-
Supplementary file 6
Tandem mass spectrometry (MS/MS) settings.
Settings used for the MS/MS analysis are presented.
- https://cdn.elifesciences.org/articles/99650/elife-99650-supp6-v2.xlsx
-
Supplementary file 7
Proteome Discoverer 2.2 settings.
Proteome Discoverer 2.2 settings for proteomics analysis are presented.
- https://cdn.elifesciences.org/articles/99650/elife-99650-supp7-v2.xlsx
-
MDAR checklist
- https://cdn.elifesciences.org/articles/99650/elife-99650-mdarchecklist1-v2.docx





