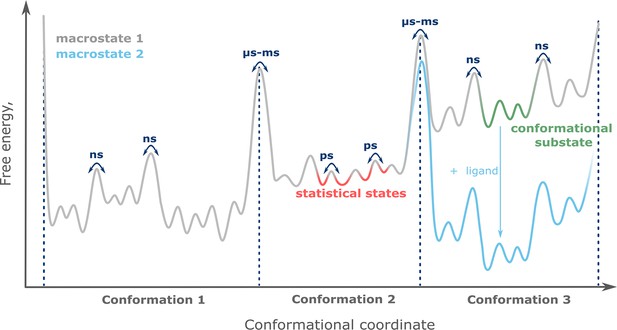
Exploring protein structural ensembles: Integration of sparse experimental data from electron paramagnetic resonance spectroscopy with molecular modeling methods
Figures
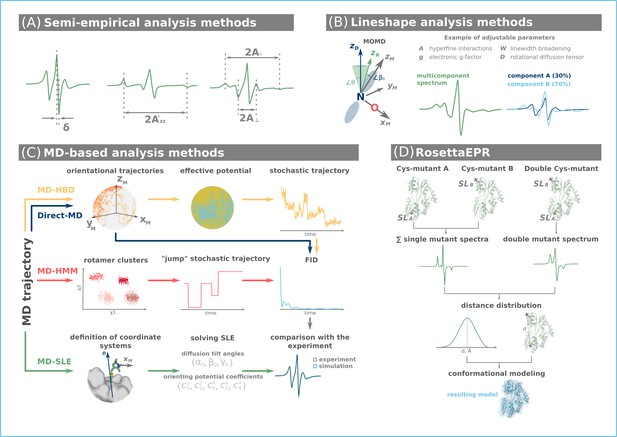
Computational approaches for the analysis and interpretations of continuous wave (CW) electron paramagnetic resonance (EPR) data.
(A) Semi-empirical analysis of the CW EPR lineshape provides insight into the rate of motion (δ), polarity (A’zz) of the spin label environment, parallel (A॥), and perpendicular (A⊥) compounds of CW EPR spectra of spin-labeled membrane bilayer lipids. (B) Lineshape analysis provides access to motional parameters and populations of individual equilibrium conformations. (C) Molecular dynamics (MD) simulations may explore the entire conformational landscape of a protein and provide data to simulate the CW EPR spectrum. The latter is then compared with the experiment. (D) RosettaEPR approach uses results from single and double mutant CW EPR experiments to derive distance constraints for subsequent conformational modeling.
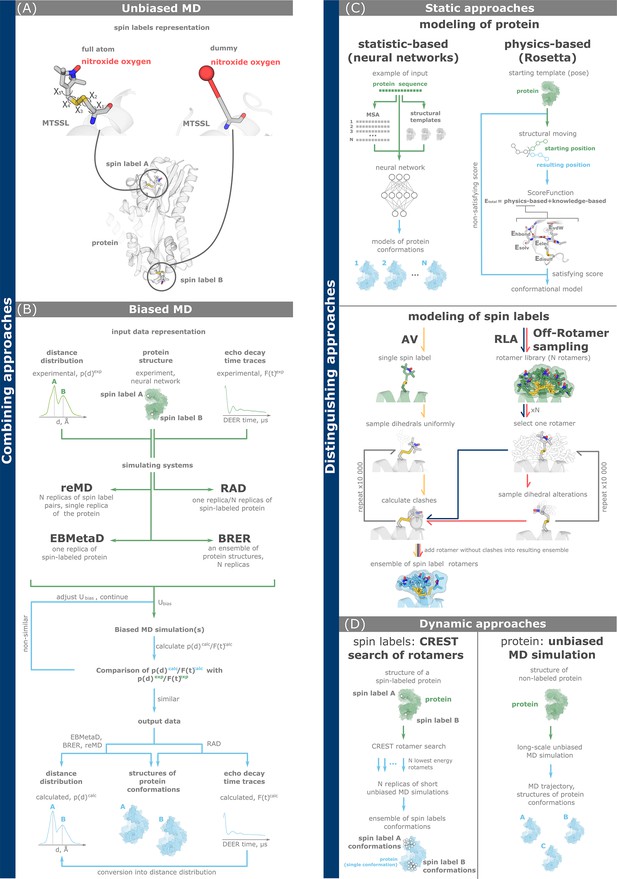
Computational methods for double electron–electron resonance (DEER) data analysis and integration.
(A, B) Combining approaches simultaneously model the dynamics of both a protein and spin labels. Full-atom and dummy representations of spin labels are possible. (C, D) Discriminating approaches investigate conformations of a protein and spin labels separately. (A) Unbiased molecular dynamics (MD) simulations of a spin-labeled protein. (B) Biased MD approaches add biasing potential to the simulating system according to the principle of maximum entropy. The potential is gradually adjusted based on the degree of agreement between simulated and experimental data, including distance distributions (reMD, EBMetaD, and BRER) and echo decay time traces (restrained average dynamics, RAD). (C) Static approaches explore the conformations of either proteins (top) using statistical and physics-based methods, or spin labels (bottom) using accessible volume (AV, yellow arrows), rotamer library approach (RLA, dark blue arrows), and off-rotamer sampling (red arrows). (D) Dynamic discriminating approaches use MD-based techniques to investigate the conformational landscapes of either spin labels (left, CREST/MD) or a protein (right, unbiased MD). In the latter case, both full-atom and coarse-grained representations of the protein are possible.
Tables
Summarizing information on electron paramagnetic resonance (EPR) spectroscopic techniques.
| Method | Features | References | |||
|---|---|---|---|---|---|
| Dynamics (timescale) | Structure(resolution) | Population | Computational analysis | ||
| CW EPR | Yes (10−10 to 10−7s) | Yes, via scanning (topology) | Yes, ≤3 conformations | Semi-empirical and lineshape analysis | Marsh, 1981; Hubbell and Altenbach, 1994; Columbus and Hubbell, 2002 |
| ST EPR | Yes (10−7 to 10−3 s) | No | No | Heuristic analysis | Hyde and Dalton, 1972 |
| TR EPR | Yes (>10−3 s) | No | Yes | Lineshape analysis | Farahbakhsh et al., 1993 |
| DEER | No | Yes (<10−10 m) | Yes | Parametric and non-parametric fitting models | Jeschke, 2012 |
| ENDOR | No | Yes (>10−11 m) | Yes | Lineshape analysis | Lubitz et al., 2002 |
| SR EPR | Yes, (10−6 to 10−5 s) | No | Yes, ≤2 conformations | Exponential fitting | Bridges et al., 2010 |