CellSeg3D, Self-supervised 3D cell segmentation for fluorescence microscopy
Figures
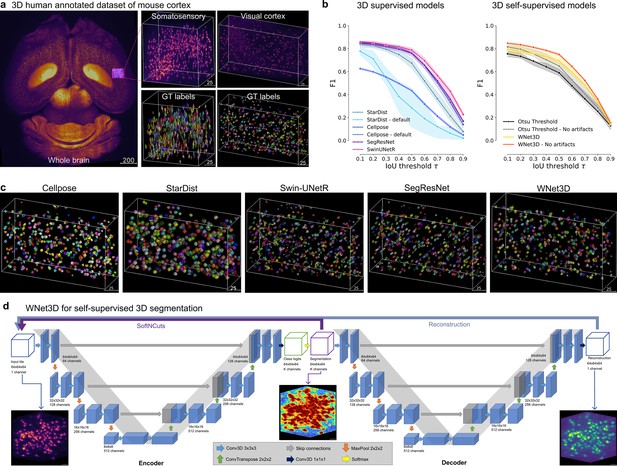
Performance of 3D semantic and instance segmentation models.
(a) Raw mesoSPIM whole-brain sample, volumes, and corresponding ground truth labels from somatosensory (S1) and visual (V1) cortical regions. (b) Evaluation of instance segmentation performance for: baseline with Otsu thresholding only, supervised models: Cellpose, StartDist, SwinUNetR, SegResNet; and our self-supervised model WNet3D over three data subsets. F1-score is computed from the Intersection over Union (IoU) with ground truth labels, then averaged. Error bars represent 50% ~Confidence Intervals (CIs). (c) View of 3D instance labels from models, as noted, for the visual cortex volume. (d) Illustration of our WNet3D architecture showcasing the dual 3D U-Net structure with our modifications (see Methods).
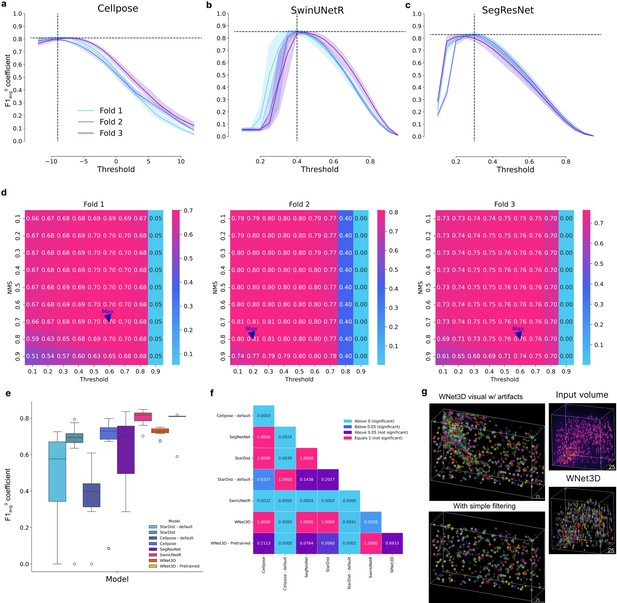
Hyperparameter tuning of baselines and statistics.
(a, b, c) Hyperparameter optimization for several supervised models. In Cellpose, the cell probability threshold value is applied before the sigmoid, hence values between −12 and 12 were tested. CellSeg3D models return predictions between 0 and 1 after applying the softmax, values tested were, therefore, in this range. Error bars show 95% ~CIs. (d) StarDist hyperparameter optimization. Several parameters were tested for non-maximum suppression (NMS) threshold and cell probability threshold. Heatmap is F1-Score. (e) Pooled F1-Scores per split, related to Figure 2a, used for statistical testing shown in f. The central box represents the interquartile range (IQR) of values with the median as a horizontal line, the upper and lower limits the upper and lower quartiles. Whiskers extend to data points within 1.5 IQR of the quartiles. Outliers are shown separately. (f) Pairwise Conover’s test p-values for the F1-Score values per model shown in e. Colors are based on level of significance. (g) Example image of WNet3D before and after artifact filtering; after also shown in Figure 1c, plus an additional example of WNet3D in S1 cortex.
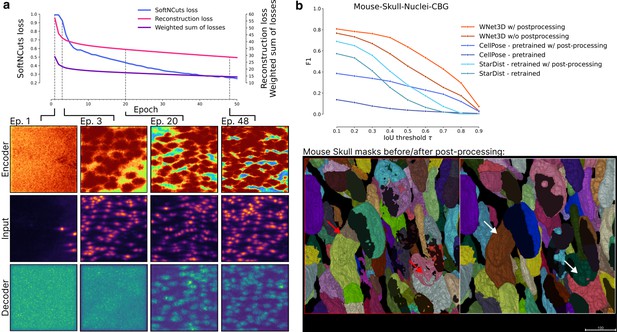
Training WNet3D: Overview of the training process of WNet3D.
(a) The loss for the encoder is the SoftNCuts, whereas the reconstruction loss for is MSE. The weighted sum of losses is calculated as indicated in Methods. For select epochs, input volumes are shown, with outputs from encoder above, and outputs from decoder below. (b) Additional model inference results on Mouse Skull dataset, and example of post-processing in order to correct holes or other artifacts, as shown with the red to white arrows.
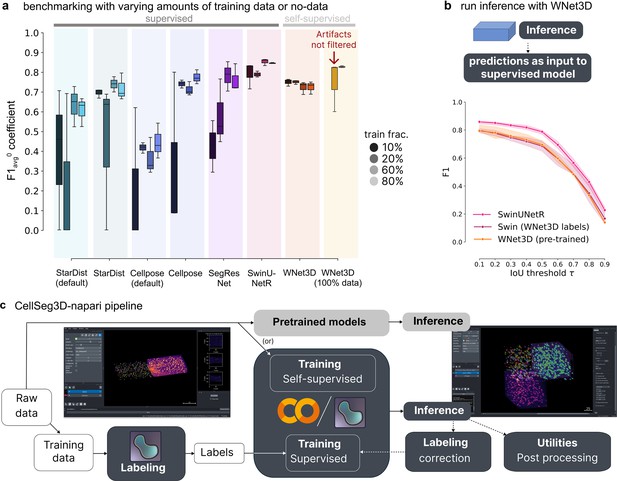
Benchmarking the performance of WNet3D vs.supervised models with various amounts of training data on our mesoSPIM dataset.
(a) Semantic segmentation performance: comparison of model efficiency, indicating the volume of training data required to achieve a given performance level. Each supervised model was trained with an increasing percentage of training data (with 10, 20, 60, or 80%, left to right/dark to light within each model grouping, see legend); F1-Score score with an was computed on unseen test data, over three data subsets for each training/evaluation split. Our self-supervised model (WNet3D) is also trained on a subset of the training set of images, but always without ground truth human labels. Far right: We also show the performance of the pre-trained WNet3D available in the plugin (far right), with and without cropping the regions where artifacts are present in the image. See Methods for details. The central box represents the interquartile range (IQR) of values with the median as a horizontal line, the upper and lower limits the upper and lower quartiles. Whiskers extend to data points within 1.5 IQR of the quartiles. (b) Instance segmentation performance comparison of Swin-UNetR and WNet3D (pretrained, see Methods), evaluated on unseen data across 3 data subsets, compared with a Swin-UNetR model trained using labels from the WNet3D self-supervised model. Here, WNet3D was trained on separate data, producing semantic labels that were then used to train a supervised Swin-UNetR model, still on held-out data. This supervised model was evaluated as the other models, on 3 held-out images from our dataset, unseen during training. Error bars indicate 50% ~CIs. (c) Workflow diagram depicting the segmentation pipeline: either raw data can be used directly (self-supervised) or labeled and used for training, after which other data can be used for model inference. Each stream concludes with post-hoc inspection and refinement, if needed (post-processing analysis and/or refining the model).
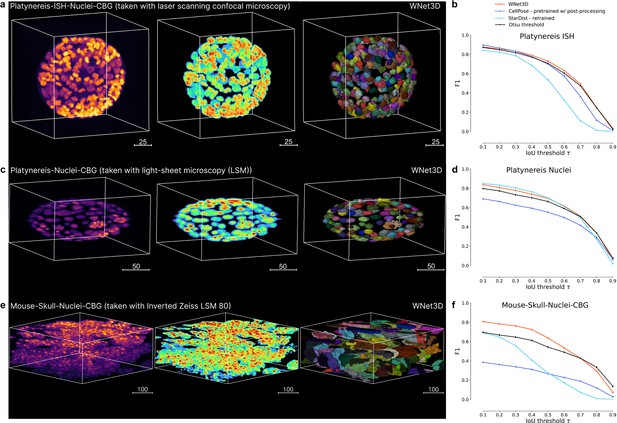
Benchmarking on additional datasets.
(a) Left: 3D Platynereis-ISH-Nuclei confocal data; middle is WNet3D semantic segmentation; right is instance segmentation. (b) Instance segmentation performance (zero-shot) of the pretrained WNet3D, Otsu threshold, and supervised models (Cellpose, StarDist) on select datasets featured in a, shown as F1-score vs intersection over union (IoU) with ground truth labels. (c) Left: 3D Platynereis-Nuclei LSM data; middle is WNet3D semantic segmentation; right is instance segmentation. (d) Instance segmentation performance (zero-shot) of the pretrained WNet3D, Otsu threshold, and supervised models (Cellpose, StarDist) on select datasets featured in c, shown as F1-score vs IoU with ground truth labels. (e) Left: Mouse Skull-Nuclei Zeiss LSM 880 data; middle is WNet3D semantic segmentation; right is instance segmentation. A demo of using CellSeg3D to obtain these results is available here: https://www.youtube.com/watch?v=U2a9IbiO7nE&t=12s. (f) Instance segmentation performance (zero-shot) of the pretrained WNet3D, Otsu threshold, and supervised models (Cellpose, StarDist) on select datasets featured in e, shown as F1-score vs IoU with ground truth labels.
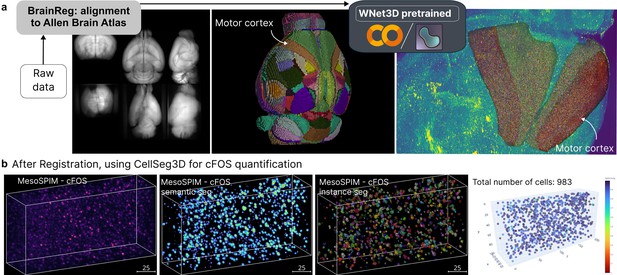
CellSeg3D napari plugin example outputs.
(a) Demo using a cleared and MesoSPIM imaged c-FOS mouse brain, followed by BrainReg (20.22) registration to the Allen Brain Atlas https://mouse.brain-map.org/, then processing of regions of interest (ROIs) with CellSeg3D. Here, the WNet3D was used for semantic segmentation followed by processing for instance segmentation. (b) Qualitative example of WNet3D-generated prediction (thresholded) and labels on a crop from the c-FOS-labeled whole-brain. A demo of using CellSeg3D to obtain these results is available here: https://www.youtube.com/watch?v=3UOvvpKxEAo.
Tables
F1-Scores for additional benchmark datasets, where we test our pretrained WNet3D, zero-shot.
Kruskal-Wallis H test [dataset, statistic, p-value]: Platynereis-ISH-Nuclei-CBG, 1.6, 0.69; Platynereis-Nuclei-CBG, 3.06, 0.38; Mouse-Skull-Nuclei-CBG (within post-processed), 10.13, 0.018; Mouse-Skull-Nuclei-CBG (no processing), 15.8, 0.001.
| F10.1 | F10.2 | F10.3 | F10.4 | F10.5 | F10.6 | F10.7 | F10.8 | F10.9 | F1MEAN | |
|---|---|---|---|---|---|---|---|---|---|---|
| Platynereis-ISH-Nuclei-CBG: | ||||||||||
| Otsu threshold | 0.872 | 0.847 | 0.817 | 0.772 | 0.706 | 0.605 | 0.474 | 0.246 | 0.026 | 0.596 |
| Cellpose (supervised) | 0.896 | 0.866 | 0.832 | 0.778 | 0.698 | 0.576 | 0.362 | 0.117 | 0.010 | 0.570 |
| StarDist (supervised) | 0.841 | 0.822 | 0.786 | 0.686 | 0.536 | 0.326 | 0.110 | 0.011 | 0. | 0.458 |
| WNet3D (zero-shot) | 0.876 | 0.856 | 0.834 | 0.790 | 0.729 | 0.632 | 0.492 | 0.249 | 0.034 | 0.610 |
| Platynereis-Nuclei-CBG: | ||||||||||
| Otsu threshold | 0.798 | 0.773 | 0.733 | 0.702 | 0.663 | 0.590 | 0.507 | 0.336 | 0.077 | 0.576 |
| Cellpose (supervised) | 0.691 | 0.663 | 0.624 | 0.594 | 0.553 | 0.497 | 0.417 | 0.290 | 0.062 | 0.488 |
| StarDist (supervised) | 0.850 | 0.833 | 0.803 | 0.764 | 0.700 | 0.611 | 0.492 | 0.272 | 0.019 | 0.594 |
| WNet3D (zero-shot) | 0.838 | 0.808 | 0.778 | 0.739 | 0.695 | 0.617 | 0.512 | 0.338 | 0.059 | 0.598 |
| Mouse-Skull-Nuclei-CBG (most challenging dataset) | ||||||||||
| Otsu threshold | 0.667 | 0.634 | 0.596 | 0.566 | 0.495 | 0.427 | 0.369 | 0.276 | 0.097 | 0.458 |
| Otsu threshold +post-processing | 0.695 | 0.668 | 0.647 | 0.612 | 0.543 | 0.490 | 0.428 | 0.334 | 0.137 | 0.506 |
| Cellpose (supervised) | 0.137 | 0.111 | 0.077 | 0.054 | 0.038 | 0.028 | 0.020 | 0.014 | 0.006 | 0.054 |
| Cellpose +post-processing | 0.386 | 0.362 | 0.339 | 0.312 | 0.266 | 0.228 | 0.189 | 0.120 | 0.027 | 0.248 |
| StarDist (supervised) | 0.573 | 0.533 | 0.411 | 0.253 | 0.135 | 0.065 | 0.020 | 0.003 | 0.0 | 0.221 |
| StarDist +post-processing | 0.689 | 0.649 | 0.557 | 0.407 | 0.276 | 0.174 | 0.073 | 0.010 | 0.0 | 0.315 |
| WNet3D (zero-shot) | 0.766 | 0.732 | 0.669 | 0.572 | 0.455 | 0.355 | 0.254 | 0.175 | 0.033 | 0.446 |
| WNet3D+post-processing | 0.807 | 0.783 | 0.763 | 0.725 | 0.637 | 0.534 | 0.428 | 0.296 | 0.073 | 0.561 |
Dataset ground-truth cell count per volume.
| Region | Size | Count |
|---|---|---|
| (pixels) | (# of cells) | |
| Sensorimotor | ||
| 1 | 199 × 106 × 147 | 343 |
| 2 | 299 × 78 × 111 | 365 |
| 3 | 299 × 105 × 147 | 631 |
| 4 | 249 × 93 × 114 | 396 |
| 5 | 249 × 86 × 94 | 347 |
| Visual | 329 × 127 × 214 | 485 |
Parameters used for instance segmentation with the pyclEsperanto Voronoi-Otsu function.
| Dataset | Outline σ | Spot σ |
|---|---|---|
| mesoSPIM | 0.65 | 0.65 |
| Mouse Skull | 1 | 15 |
| Platynereis-ISH | 0.5 | 2 |
| Platynereis | 0.5 | 2.75 |





