Wavenumber-dependent transmission of subthreshold waves on electrical synapses network model of Caenorhabditis elegans
Figures
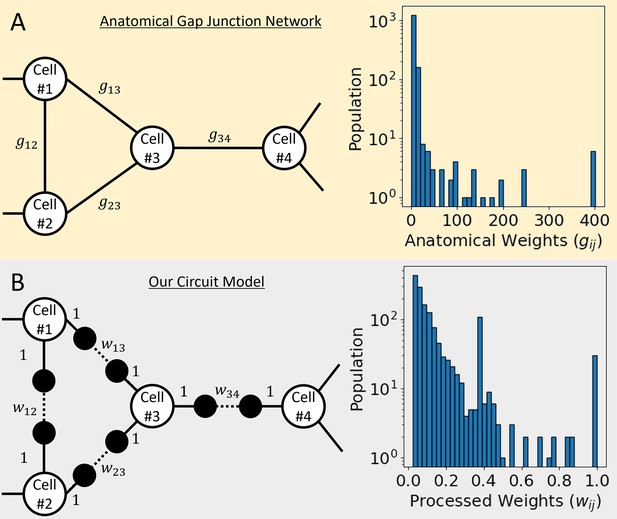
Anatomical gap junction network and our circuit model.
(A) In the sample of the network diagram on the left, there are white-filled nodes stand for cells, and the edges of a solid line stand for the anatomical gap junction between them. The graph on the right is the distribution of 1433 gap junction weights obtained from the connectome data of C. elegans. (B) In the sample of the network diagram on the left, equally spaced two virtual nodes were added between cells connected by a gap junction, and the virtual node is presented as a black-filled circle. The solid line stands for the connection between the cell and the virtual node that gave a weight of 1, and the dotted line stands for the connection between the two virtual nodes that gave our processed weight between 0 and 1. Our processed weights are calculated from the anatomical gap junction weights, and the graph on the right shows its distribution.
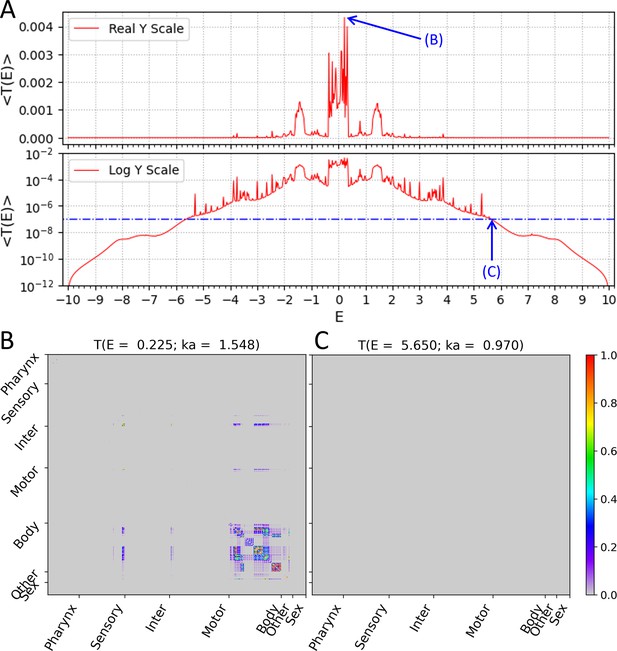
Wavenumber-dependent transmission coefficient.
Average transmission coefficient for all cell-pairs as a function of E values (E value is a function of the wavenumber of the subthreshold wave). (A) The upper panel shows the average transmission coefficient graph on the Y-axis of the real scale and the lower panel shows the same graph on the Y-axis of the log scale. The reference value (10–7) of the signal mobility edge is indicated in the lower panel as a dot-dash line. (B) The transmission coefficient of all cell-pairs at is represented as a heatmap, which is the highest average transmission coefficient in our result. (C) The transmission coefficient of all cell-pairs at is represented as a heatmap, which is one of the signal mobility edges. (B, C) In heatmap, the X and Y axes are arranged according to the index of 469 cells, and the index follows that of the original connectome data. Instead of displaying all cell names, only seven cell types were indicated.
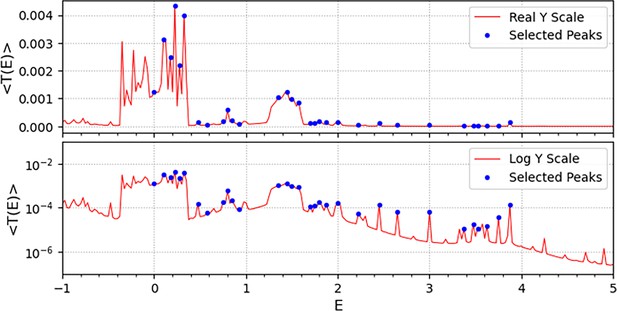
Selected peaks.
The 31 selected peaks are shown as dots on the graph of the average transmission coefficient for all cell-pairs. To make the dots more visible, only the E value range of [–1, 5] was drawn out of the total [–10, 10]. The upper panel shows the dots and the graph on the Y-axis of the real scale and the lower panel shows the same things on the Y-axis of the log scale.
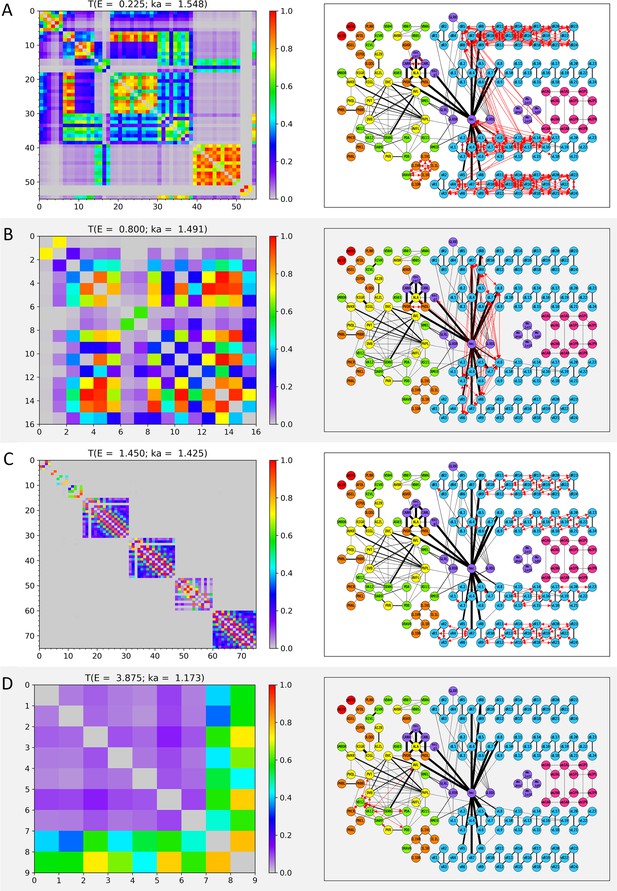
Wavenumber-dependent transmission map.
The cell-pairs with strong transmission () at (A) , (B) , (C) , and (D) (or the corresponding wavenumbers) are shown, respectively. The left panels represent the transmission coefficient between cells belonging to the cell-pairs with strong transmission at each E value as a heatmap. Here, the X and Y axes are the cell index, and the cell name corresponding to the cell index can be found in Appendix 1—table 1, Appendix 1—table 2, Appendix 1—table 3, Appendix 1—table 4. The right panels display the cell-pairs of strong transmission at each E value as red bidirectional arrows of the same thickness on the network diagram. In the network diagram, the cell name is written inside the circle that stands for each node (for the body-wall muscles, the cell name is abbreviated as follows: ex) dBWML1 → dL1, and the color of the node stands for the cell type (red: pharynx cells, orange: sensory neurons, yellow: inter neurons, green: motor neurons, light blue: body-wall muscles, purple: other end organs, magenta: sex-specific cells). Edges indicated by black solid lines stand for the electrical synapses among the cells, and the thickness of the edges is proportional to our processed weight. In the network diagram, only 173 cells that belonged to the cell-pair with strong transmission at least once in positive E values are represented, and the remaining cells and the electrical synapses by them are omitted from display. The virtual nodes of our circuit are also omitted from the display.
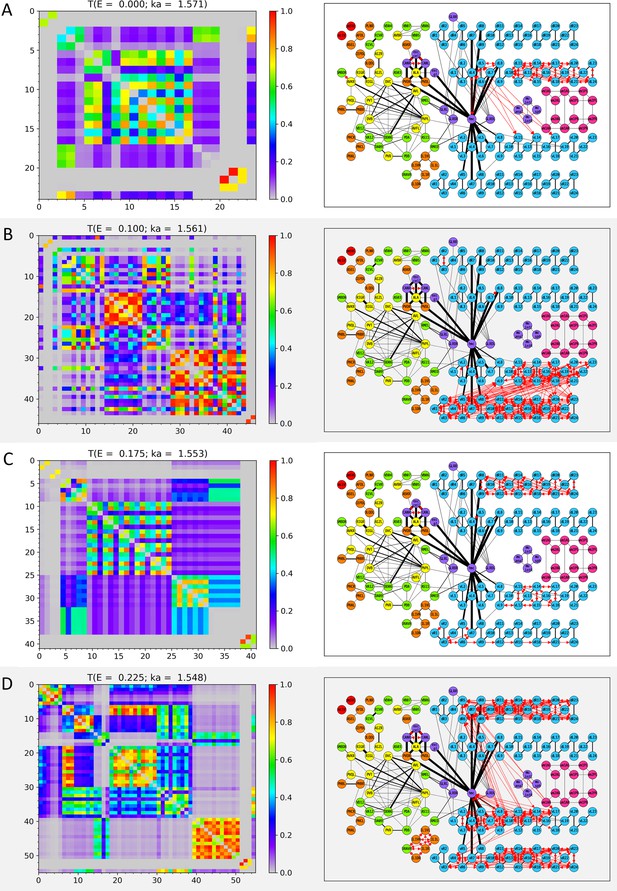
Wavenumber-dependent transmission map.
The cell-pairs with strong transmission () at (A) , (B) , (C) , and (D) (or the corresponding wavenumbers) are shown, respectively. The left panels represent the transmission coefficient between cells belonging to the cell-pairs with strong transmission at each E value as a heatmap. Here, the X and Y axes are the cell index, and the cell name corresponding to the cell index can be found in Appendix 1—Tables 1–4. The right panels display the cell-pairs of strong transmission at each E value as red bidirectional arrows of the same thickness on the network diagram. In the network diagram, the cell name is written inside the circle that stands for each node (for the body-wall muscles, the cell name is abbreviated as follows: ex) dBWML1 → dL1, and the color of the node stands for the cell type (red: pharynx cells, orange: sensory neurons, yellow: inter neurons, green: motor neurons, light blue: body-wall muscles, purple: other end organs, magenta: sex-specific cells). Edges indicated by black solid lines stand for the electrical synapses among the cells, and the thickness of the edges is proportional to our processed weight. In the network diagram, only 173 cells that belonged to the cell-pair with strong transmission at least once in positive E values are represented, and the remaining cells and the electrical synapses by them are omitted from display. The virtual nodes of our circuit are also omitted from the display.
Wavenumber-dependent transmission map.
The cell-pairs with strong transmission () at (A) , (B) , (C) , and (D) (or the corresponding wavenumbers) are shown, respectively. The heatmaps of the left panels and the network diagram of the right panels are represented in the same way as in Figure 3—figure supplement 1.
Wavenumber-dependent transmission map.
The cell-pairs with strong transmission () at (A) , (B) , (C) , and (D) (or the corresponding wavenumbers) are shown, respectively. The heatmaps of the left panels and the network diagram of the right panels are represented in the same way as in Figure 3—figure supplement 1.
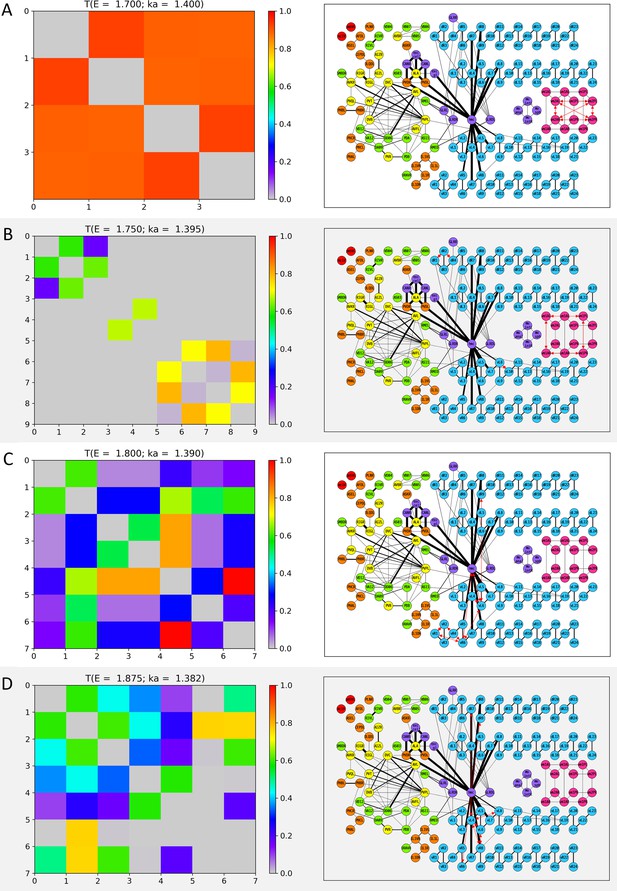
Wavenumber-dependent transmission map.
The cell-pairs with strong transmission () at (A) , (B) , (C) , and (D) (or the corresponding wavenumbers) are shown, respectively. The heatmaps of the left panels and the network diagram of the right panels are represented in the same way as in Figure 3—figure supplement 1.
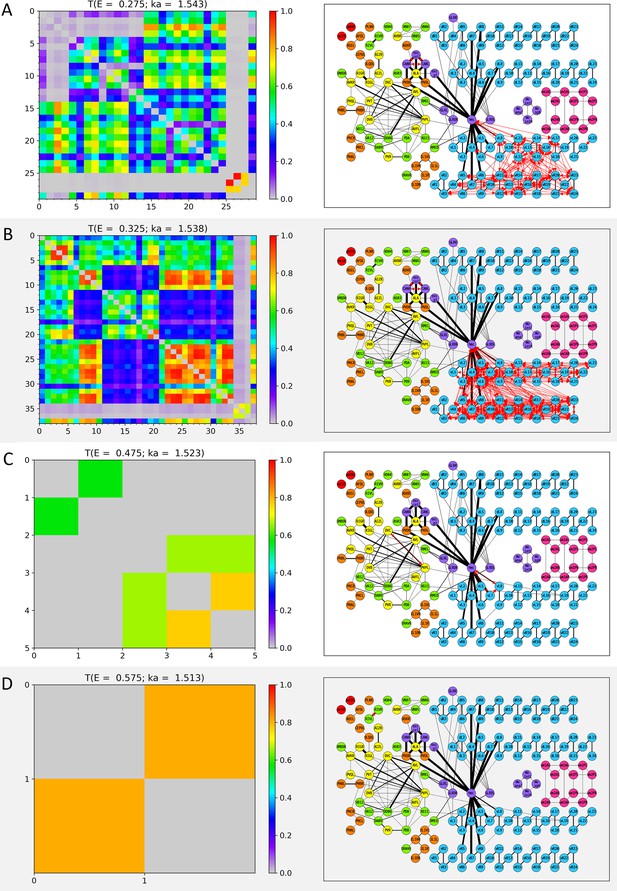
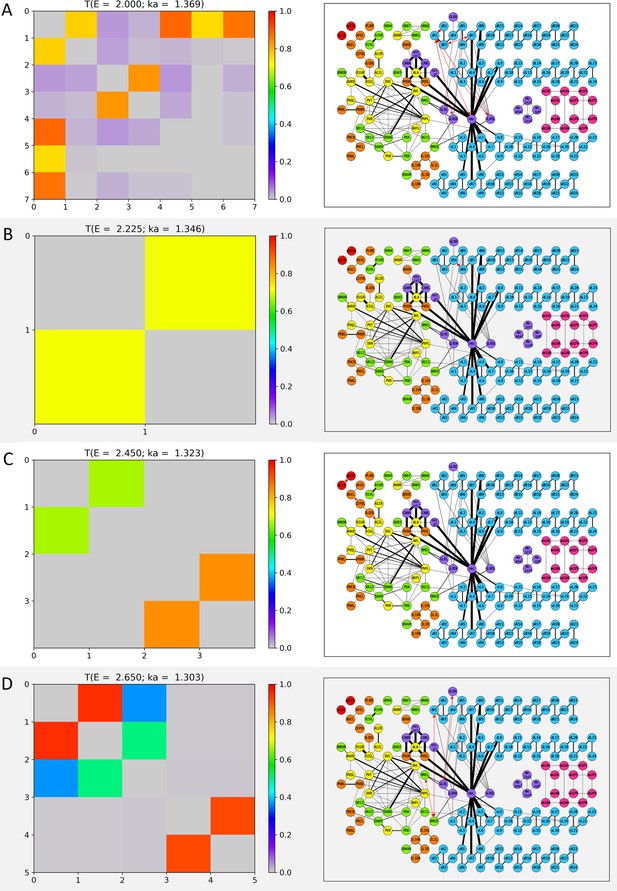
Wavenumber-dependent transmission map.
The cell-pairs with strong transmission () at (A) , (B) , (C) , and (D) (or the corresponding wavenumbers) are shown, respectively. The heatmaps of the left panels and the network diagram of the right panels are represented in the same way as in Figure 3—figure supplement 1.
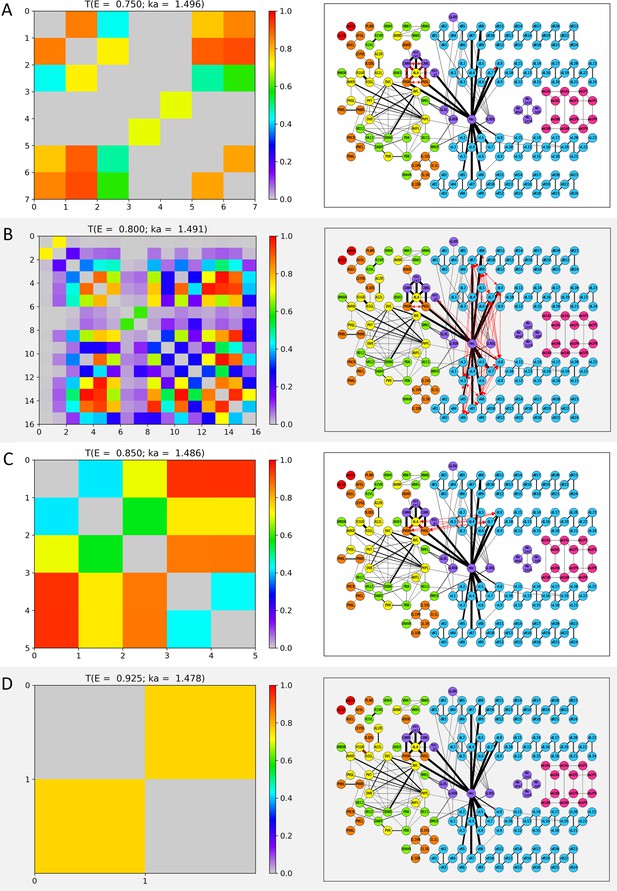
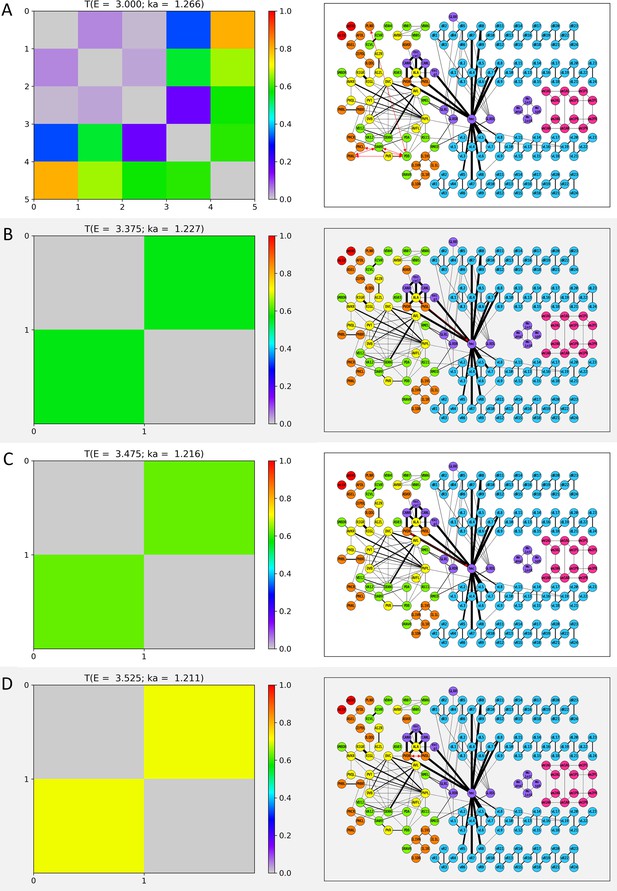
Wavenumber-dependent transmission map.
The cell-pairs with strong transmission () at (A) , (B) , (C) , and (D) (or the corresponding wavenumbers) are shown, respectively. The heatmaps of the left panels and the network diagram of the right panels are represented in the same way as in Figure 3—figure supplement 1.
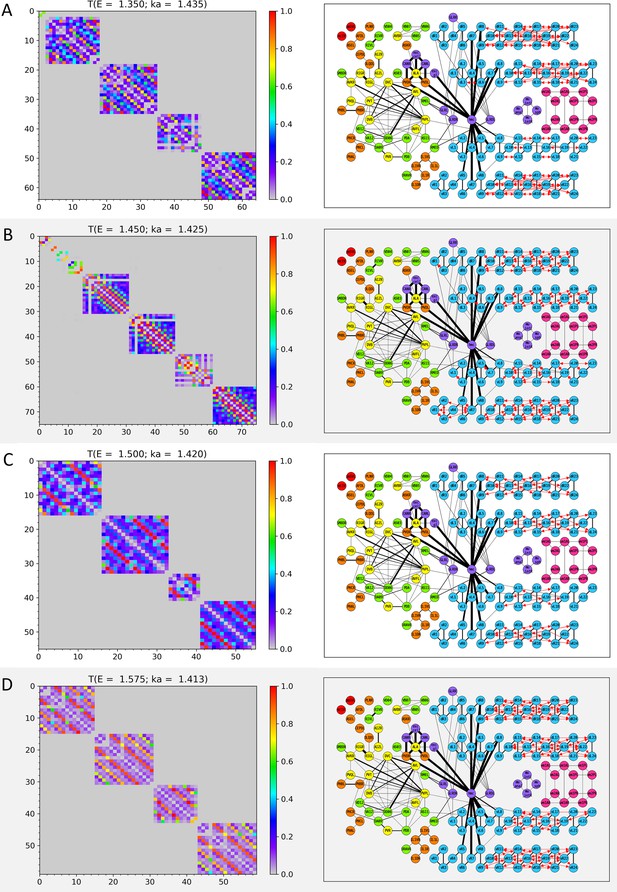
Wavenumber-dependent transmission map.
The cell-pairs with strong transmission () at (A) , (B) , (C) , and (D) (or the corresponding wavenumbers) are shown, respectively. The heatmaps of the left panels and the network diagram of the right panels are represented in the same way as in Figure 3—figure supplement 1.
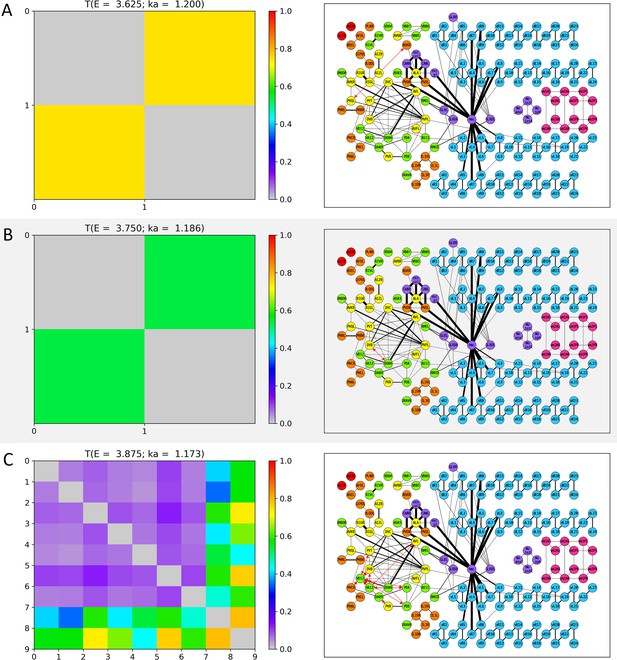
Wavenumber-dependent transmission map.
The cell-pairs with strong transmission () at (A) , (B) , and (C) (or the corresponding wavenumbers) are shown, respectively. The heatmaps of the left panels and the network diagram of the right panels are represented in the same way as in Figure 3—figure supplement 1.
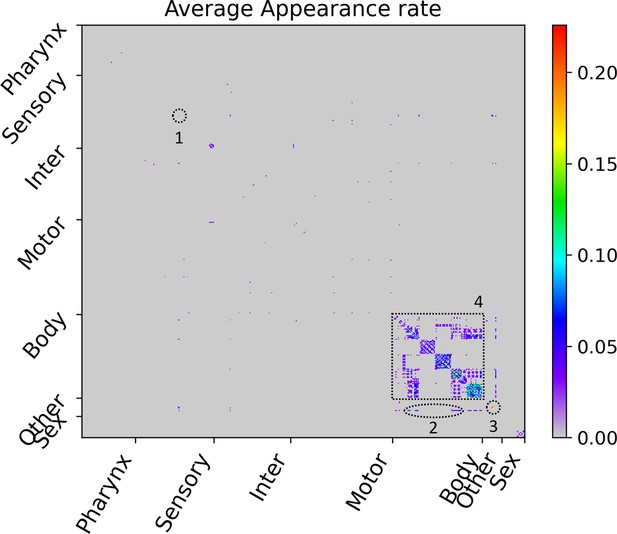
Average appearance rate as cell-pair with strong transmission in the WDTMs.
The average appearance rate of all cell-pairs is represented as a heatmap. Here, the X and Y axes are arranged according to the index of 469 cells, and the index follows that of the original connectome data. Instead of displaying all cell names, only seven cell types were indicated. The highest rate is , and four cell-pair groups showing rates of or higher are identified and numbered in the heatmap. (1) PVDL-PVDR pair, (2) hmc-vBWML6, hmc-vBWML7, and hmc-vBWML8 pairs, (3) CANL-CANR, CANL-exc_cell, and CANR-exc_cell pairs, and (4) many cell-pairs of intra-strand of the body-wall muscles.
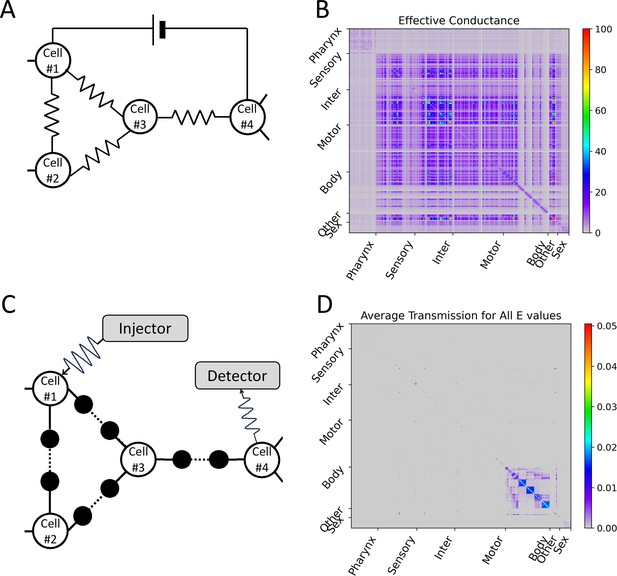
Differences depending on whether the electrical synapse network model considers interference.
(A) In the electrical synapse network model without considering interference, when the electrical synapse is treated as an electrical resistor, the effective conductance (the reciprocal of the effective resistance) experienced by the external direct current power applied to an arbitrary cell-pair was calculated, and this value was represented as (B) a heatmap for all cell-pairs. (C) In our circuit considering interference of wave signals, the average transmission coefficient for all E values (or all wavenumbers) of an arbitrary cell-pair was calculated, and this value was represented as (D) a heatmap for all cell-pairs. (B, D) In heatmap, the X and Y axes are arranged according to the index of 469 cells, and the index follows that of the original connectome data. Instead of displaying all cell names, only seven cell types are indicated.
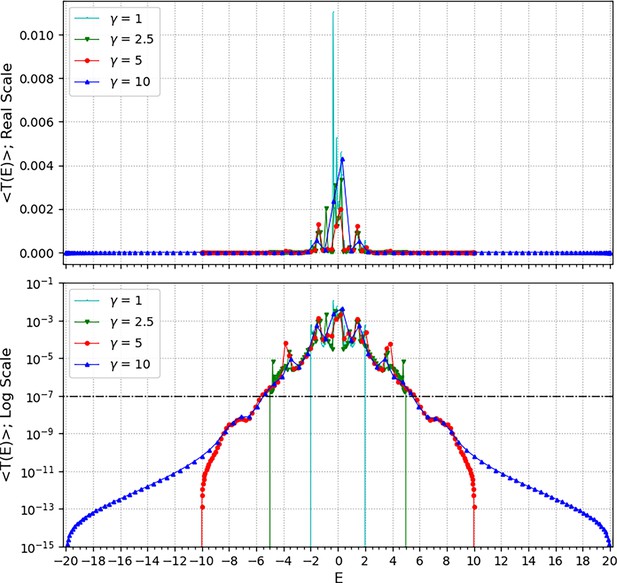
Searching for optimal gamma.
The average transmission coefficient for all cell-pairs as a function of E value (E value is a function of both and wavenumber k) was calculated under the four conditions (1, 2.5, 5, and 10), respectively. Here, the wavenumbers used the same set as a hundred values equally spaced in the range of [0, ]. The upper panel shows the average transmission coefficient graphs as the Y-axis of the real scale and the lower panel shows the same graphs as the Y-axis of the log scale. The reference value (10–7) of the signal mobility edge is shown in the lower panel as a dot-dash line.
Tables
Cell names corresponding to the indices in the heatmaps for wavenumber-dependent transmission map.
Each two columns shows the index and the corresponding cell name of the cells that belonged to the cell-pairs with strong transmission at the E value (or the corresponding wavenumber) indicated in the first row. The color painted on the name box stands for the cell type (red: pharynx cells, orange: sensory neurons, yellow: inter neurons, green: motor neurons, light blue: body-wall muscles, purple: other end organs, magenta: sex-specific cells).
| E = 0.000 | ka = 1.571 | E = 0.100 | ka = 1.561 | E = 0.175 | ka = 1.553 | E = 0.225 | ka = 1.548 | E = 0.275 | ka = 1.543 | E = 0.325 | ka = 1.538 | E = 0.475 | ka = 1.523 | E = 0.575 | ka = 1.513 |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Index | Cell name | Index | Cell name | Index | Cell name | Index | Cell name | Index | Cell name | Index | Cell name | Index | Cell name | Index | Cell name |
| 0 | AVKR | 0 | dBWML7 | 0 | dBWML4 | 0 | IL1DR | 0 | vBWML4 | 0 | vBWML1 | 0 | PVPL | 0 | RIVL |
| 1 | SMBDR | 1 | dBWMR2 | 1 | dBWML5 | 1 | IL1L | 1 | vBWMR3 | 1 | vBWML3 | 1 | DVC | 1 | RIVR |
| 2 | dBWML4 | 2 | dBWMR3 | 2 | vBWMR1 | 2 | IL1R | 2 | vBWMR4 | 2 | vBWML4 | 2 | vBWML7 | ||
| 3 | dBWML5 | 3 | vBWML2 | 3 | vBWMR2 | 3 | IL1VL | 3 | vBWMR5 | 3 | vBWML5 | 3 | vBWML8 | ||
| 4 | dBWML8 | 4 | vBWML6 | 4 | vBWMR3 | 4 | IL1VR | 4 | vBWML8 | 4 | vBWML6 | 4 | hmc | ||
| 5 | dBWML10 | 5 | vBWMR1 | 5 | vBWMR4 | 5 | URAVR | 5 | vBWML10 | 5 | vBWML7 | ||||
| 6 | dBWML11 | 6 | vBWMR2 | 6 | vBWMR5 | 6 | dBWMR5 | 6 | vBWML11 | 6 | vBWMR3 | ||||
| 7 | dBWML12 | 7 | vBWMR3 | 7 | vBWMR6 | 7 | dBWMR6 | 7 | vBWML12 | 7 | vBWMR4 | ||||
| 8 | dBWML13 | 8 | vBWMR4 | 8 | vBWMR7 | 8 | dBWMR7 | 8 | vBWML13 | 8 | vBWMR5 | ||||
| 9 | dBWML14 | 9 | vBWMR5 | 9 | dBWMR8 | 9 | vBWML2 | 9 | vBWML15 | 9 | vBWMR6 | ||||
| 10 | dBWML15 | 10 | vBWMR6 | 10 | dBWMR9 | 10 | vBWML3 | 10 | vBWML16 | 10 | vBWMR7 | ||||
| 11 | dBWML17 | 11 | vBWMR7 | 11 | dBWMR10 | 11 | vBWML4 | 11 | vBWML17 | 11 | vBWML10 | ||||
| 12 | dBWML18 | 12 | dBWML8 | 12 | dBWMR11 | 12 | vBWML6 | 12 | vBWML22 | 12 | vBWML11 | ||||
| 13 | dBWML20 | 13 | vBWML9 | 13 | dBWMR12 | 13 | vBWML7 | 13 | vBWML23 | 13 | vBWML12 | ||||
| 14 | dBWML21 | 14 | vBWML10 | 14 | dBWMR13 | 14 | vBWMR3 | 14 | vBWMR10 | 14 | vBWML16 | ||||
| 15 | dBWML23 | 15 | vBWML11 | 15 | dBWMR14 | 15 | vBWMR4 | 15 | vBWMR11 | 15 | vBWML17 | ||||
| 16 | dBWML24 | 16 | vBWML12 | 16 | dBWMR15 | 16 | vBWMR6 | 16 | vBWMR12 | 16 | vBWML18 | ||||
| 17 | vBWML11 | 17 | vBWML13 | 17 | dBWMR16 | 17 | vBWMR7 | 17 | vBWMR14 | 17 | vBWML19 | ||||
| 18 | vBWML14 | 18 | vBWML15 | 18 | dBWMR17 | 18 | dBWMR9 | 18 | vBWMR15 | 18 | vBWML21 | ||||
| 19 | vBWML17 | 19 | vBWML16 | 19 | dBWMR18 | 19 | dBWMR10 | 19 | vBWMR16 | 19 | vBWML22 | ||||
| 20 | CANL | 20 | vBWML17 | 20 | dBWMR19 | 20 | dBWMR11 | 20 | vBWMR19 | 20 | vBWML23 | ||||
| 21 | CANR | 21 | vBWML18 | 21 | dBWMR20 | 21 | dBWMR13 | 21 | vBWMR20 | 21 | vBWMR9 | ||||
| 22 | exc_cell | 22 | vBWML19 | 22 | dBWMR21 | 22 | dBWMR14 | 22 | vBWMR21 | 22 | vBWMR10 | ||||
| 23 | hmc | 23 | vBWML20 | 23 | dBWMR23 | 23 | dBWMR16 | 23 | vBWMR23 | 23 | vBWMR11 | ||||
| 24 | vBWML21 | 24 | dBWMR24 | 24 | dBWMR17 | 24 | vBWMR24 | 24 | vBWMR12 | ||||||
| 25 | vBWML22 | 25 | vBWML9 | 25 | dBWMR18 | 25 | CANL | 25 | vBWMR13 | ||||||
| 26 | vBWML23 | 26 | vBWML11 | 26 | dBWMR20 | 26 | CANR | 26 | vBWMR16 | ||||||
| 27 | vBWMR8 | 27 | vBWML12 | 27 | dBWMR21 | 27 | exc_cell | 27 | vBWMR17 | ||||||
| 28 | vBWMR9 | 28 | vBWML14 | 28 | dBWMR23 | 28 | hmc | 28 | vBWMR18 | ||||||
| 29 | vBWMR10 | 29 | vBWML15 | 29 | dBWMR24 | 29 | vBWMR19 | ||||||||
| 30 | vBWMR11 | 30 | vBWML17 | 30 | vBWML8 | 30 | vBWMR20 | ||||||||
| 31 | vBWMR12 | 31 | vBWML18 | 31 | vBWML9 | 31 | vBWMR22 | ||||||||
| 32 | vBWMR13 | 32 | vBWMR9 | 32 | vBWML10 | 32 | vBWMR23 | ||||||||
| 33 | vBWMR14 | 33 | vBWMR12 | 33 | vBWML11 | 33 | vBWMR24 | ||||||||
| 34 | vBWMR15 | 34 | vBWMR15 | 34 | vBWML13 | 34 | CANL | ||||||||
| 35 | vBWMR16 | 35 | vBWMR18 | 35 | vBWML14 | 35 | CANR | ||||||||
| 36 | vBWMR17 | 36 | vBWMR21 | 36 | vBWML16 | 36 | exc_cell | ||||||||
| 37 | vBWMR18 | 37 | vBWMR24 | 37 | vBWML17 | 37 | hmc | ||||||||
| 38 | vBWMR19 | 38 | CANL | 38 | vBWML18 | ||||||||||
| 39 | vBWMR20 | 39 | CANR | 39 | vBWMR9 | ||||||||||
| 40 | vBWMR21 | 40 | exc_cell | 40 | vBWMR10 | ||||||||||
| 41 | vBWMR22 | 41 | vBWMR11 | ||||||||||||
| 42 | vBWMR23 | 42 | vBWMR13 | ||||||||||||
| 43 | vBWMR24 | 43 | vBWMR14 | ||||||||||||
| 44 | CANL | 44 | vBWMR16 | ||||||||||||
| 45 | CANR | 45 | vBWMR17 | ||||||||||||
| 46 | vBWMR18 | ||||||||||||||
| 47 | vBWMR20 | ||||||||||||||
| 48 | vBWMR21 | ||||||||||||||
| 49 | vBWMR23 | ||||||||||||||
| 50 | vBWMR24 | ||||||||||||||
| 51 | CANL | ||||||||||||||
| 52 | CANR | ||||||||||||||
| 53 | exc_cell | ||||||||||||||
| 54 | hmc |
Cell names corresponding to the indices in the heatmaps for wavenumber-dependent transmission map.
It is represented in the same way as Appendix 1—table 1.
| E = 0.750 | ka = 1.496 | E = 0.800 | ka = 1.491 | E = 0.850 | ka = 1.486 | E = 0.925 | ka = 1.478 | E = 1.350 | ka = 1.435 | E = 1.450 | ka = 1.425 | E = 1.500 | ka = 1.420 | E = 1.575 | ka = 1.413 |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Index | Cell name | Index | Cell name | Index | Cell name | Index | Cell name | Index | Cell name | Index | Cell name | Index | Cell name | Index | Cell name |
| 0 | PVDL | 0 | PVDL | 0 | PVDL | 0 | RIGL | 0 | dBWML3 | 0 | dBWML3 | 0 | dBWML9 | 0 | dBWML10 |
| 1 | PVDR | 1 | PVDR | 1 | PVDR | 1 | RIGR | 1 | dBWML6 | 1 | dBWML4 | 1 | dBWML10 | 1 | dBWML11 |
| 2 | ALA | 2 | ALA | 2 | ALA | 2 | dBWML9 | 2 | dBWML5 | 2 | dBWML11 | 2 | dBWML12 | ||
| 3 | dBWML7 | 3 | dBWML7 | 3 | dBWML7 | 3 | dBWML10 | 3 | dBWML6 | 3 | dBWML12 | 3 | dBWML13 | ||
| 4 | dBWML8 | 4 | dBWMR7 | 4 | dBWML8 | 4 | dBWML11 | 4 | dBWMR1 | 4 | dBWML13 | 4 | dBWML14 | ||
| 5 | CANL | 5 | vBWML3 | 5 | dBWML12 | 5 | dBWMR3 | 5 | dBWML14 | 5 | dBWML15 | ||||
| 6 | CANR | 6 | vBWML5 | 6 | dBWML13 | 6 | vBWML1 | 6 | dBWML15 | 6 | dBWML16 | ||||
| 7 | vBWML7 | 7 | dBWML14 | 7 | vBWML3 | 7 | dBWML16 | 7 | dBWML17 | ||||||
| 8 | vBWMR6 | 8 | dBWML15 | 8 | vBWML5 | 8 | dBWML17 | 8 | dBWML18 | ||||||
| 9 | vBWMR7 | 9 | dBWML16 | 9 | vBWML7 | 9 | dBWML18 | 9 | dBWML19 | ||||||
| 10 | dBWML8 | 10 | dBWML17 | 10 | vBWMR1 | 10 | dBWML19 | 10 | dBWML20 | ||||||
| 11 | dBWMR8 | 11 | dBWML18 | 11 | vBWMR2 | 11 | dBWML20 | 11 | dBWML21 | ||||||
| 12 | dBWMR9 | 12 | dBWML19 | 12 | vBWMR3 | 12 | dBWML21 | 12 | dBWML22 | ||||||
| 13 | vBWML8 | 13 | dBWML20 | 13 | vBWMR5 | 13 | dBWML22 | 13 | dBWML23 | ||||||
| 14 | vBWMR8 | 14 | dBWML21 | 14 | vBWMR7 | 14 | dBWML23 | 14 | dBWML24 | ||||||
| 15 | exc_gl | 15 | dBWML22 | 15 | dBWML9 | 15 | dBWML24 | 15 | dBWMR9 | ||||||
| 16 | dBWML23 | 16 | dBWML10 | 16 | dBWMR8 | 16 | dBWMR10 | ||||||||
| 17 | dBWML24 | 17 | dBWML11 | 17 | dBWMR9 | 17 | dBWMR11 | ||||||||
| 18 | dBWMR8 | 18 | dBWML12 | 18 | dBWMR10 | 18 | dBWMR12 | ||||||||
| 19 | dBWMR9 | 19 | dBWML13 | 19 | dBWMR11 | 19 | dBWMR13 | ||||||||
| 20 | dBWMR10 | 20 | dBWML14 | 20 | dBWMR12 | 20 | dBWMR14 | ||||||||
| 21 | dBWMR11 | 21 | dBWML15 | 21 | dBWMR13 | 21 | dBWMR15 | ||||||||
| 22 | dBWMR12 | 22 | dBWML16 | 22 | dBWMR14 | 22 | dBWMR16 | ||||||||
| 23 | dBWMR13 | 23 | dBWML17 | 23 | dBWMR15 | 23 | dBWMR17 | ||||||||
| 24 | dBWMR14 | 24 | dBWML18 | 24 | dBWMR16 | 24 | dBWMR18 | ||||||||
| 25 | dBWMR15 | 25 | dBWML19 | 25 | dBWMR17 | 25 | dBWMR19 | ||||||||
| 26 | dBWMR16 | 26 | dBWML20 | 26 | dBWMR18 | 26 | dBWMR20 | ||||||||
| 27 | dBWMR17 | 27 | dBWML21 | 27 | dBWMR19 | 27 | dBWMR21 | ||||||||
| 28 | dBWMR18 | 28 | dBWML22 | 28 | dBWMR20 | 28 | dBWMR22 | ||||||||
| 29 | dBWMR19 | 29 | dBWML23 | 29 | dBWMR21 | 29 | dBWMR23 | ||||||||
| 30 | dBWMR20 | 30 | dBWML24 | 30 | dBWMR22 | 30 | dBWMR24 | ||||||||
| 31 | dBWMR21 | 31 | dBWMR8 | 31 | dBWMR23 | 31 | vBWML9 | ||||||||
| 32 | dBWMR22 | 32 | dBWMR9 | 32 | dBWMR24 | 32 | vBWML10 | ||||||||
| 33 | dBWMR23 | 33 | dBWMR10 | 33 | vBWML9 | 33 | vBWML11 | ||||||||
| 34 | dBWMR24 | 34 | dBWMR11 | 34 | vBWML10 | 34 | vBWML12 | ||||||||
| 35 | vBWML9 | 35 | dBWMR13 | 35 | vBWML11 | 35 | vBWML13 | ||||||||
| 36 | vBWML10 | 36 | dBWMR14 | 36 | vBWML12 | 36 | vBWML14 | ||||||||
| 37 | vBWML11 | 37 | dBWMR15 | 37 | vBWML16 | 37 | vBWML15 | ||||||||
| 38 | vBWML12 | 38 | dBWMR16 | 38 | vBWML17 | 38 | vBWML16 | ||||||||
| 39 | vBWML13 | 39 | dBWMR17 | 39 | vBWML18 | 39 | vBWML17 | ||||||||
| 40 | vBWML14 | 40 | dBWMR18 | 40 | vBWML19 | 40 | vBWML18 | ||||||||
| 41 | vBWML15 | 41 | dBWMR19 | 41 | vBWMR9 | 41 | vBWML19 | ||||||||
| 42 | vBWML16 | 42 | dBWMR20 | 42 | vBWMR10 | 42 | vBWML23 | ||||||||
| 43 | vBWML17 | 43 | dBWMR21 | 43 | vBWMR11 | 43 | vBWMR9 | ||||||||
| 44 | vBWML18 | 44 | dBWMR22 | 44 | vBWMR12 | 44 | vBWMR10 | ||||||||
| 45 | vBWML20 | 45 | dBWMR23 | 45 | vBWMR13 | 45 | vBWMR11 | ||||||||
| 46 | vBWML22 | 46 | dBWMR24 | 46 | vBWMR14 | 46 | vBWMR12 | ||||||||
| 47 | vBWML23 | 47 | vBWML9 | 47 | vBWMR16 | 47 | vBWMR13 | ||||||||
| 48 | vBWMR9 | 48 | vBWML10 | 48 | vBWMR17 | 48 | vBWMR14 | ||||||||
| 49 | vBWMR10 | 49 | vBWML11 | 49 | vBWMR18 | 49 | vBWMR15 | ||||||||
| 50 | vBWMR11 | 50 | vBWML12 | 50 | vBWMR19 | 50 | vBWMR16 | ||||||||
| 51 | vBWMR12 | 51 | vBWML14 | 51 | vBWMR20 | 51 | vBWMR17 | ||||||||
| 52 | vBWMR13 | 52 | vBWML16 | 52 | vBWMR21 | 52 | vBWMR18 | ||||||||
| 53 | vBWMR14 | 53 | vBWML17 | 53 | vBWMR23 | 53 | vBWMR19 | ||||||||
| 54 | vBWMR15 | 54 | vBWML18 | 54 | vBWMR24 | 54 | vBWMR20 | ||||||||
| 55 | vBWMR16 | 55 | vBWML19 | 55 | vBWMR21 | ||||||||||
| 56 | vBWMR17 | 56 | vBWML20 | 56 | vBWMR22 | ||||||||||
| 57 | vBWMR18 | 57 | vBWML21 | 57 | vBWMR23 | ||||||||||
| 58 | vBWMR19 | 58 | vBWML22 | 58 | vBWMR24 | ||||||||||
| 59 | vBWMR20 | 59 | vBWML23 | ||||||||||||
| 60 | vBWMR21 | 60 | vBWMR9 | ||||||||||||
| 61 | vBWMR22 | 61 | vBWMR11 | ||||||||||||
| 62 | vBWMR23 | 62 | vBWMR12 | ||||||||||||
| 63 | vBWMR24 | 63 | vBWMR13 | ||||||||||||
| 64 | vBWMR14 | ||||||||||||||
| 65 | vBWMR15 | ||||||||||||||
| 66 | vBWMR16 | ||||||||||||||
| 67 | vBWMR17 | ||||||||||||||
| 68 | vBWMR18 | ||||||||||||||
| 69 | vBWMR19 | ||||||||||||||
| 70 | vBWMR20 | ||||||||||||||
| 71 | vBWMR21 | ||||||||||||||
| 72 | vBWMR22 | ||||||||||||||
| 73 | vBWMR23 | ||||||||||||||
| 74 | vBWMR24 |
Cell names corresponding to the indices in the heatmaps for wavenumber-dependent transmission map.
It is represented in the same way as Appendix 1—table 1.
| E = 1.700 | ka = 1.400 | E = 1.750 | ka = 1.395 | E = 1.800 | ka = 1.390 | E = 1.875 | ka = 1.382 | E = 2.000 | ka = 1.369 | E = 2.225 | ka = 1.346 | E = 2.450 | ka = 1.323 | E = 2.650 | ka = 1.303 |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Index | Cell name | Index | Cell name | Index | Cell name | Index | Cell name | Index | Cell name | Index | Cell name | Index | Cell name | Index | Cell name |
| 0 | vm2AL | 0 | dBWMR1 | 0 | vBWML4 | 0 | dBWMR7 | 0 | dBWMR1 | 0 | dBWMR4 | 0 | pm5VL | 0 | RMEL |
| 1 | vm2AR | 1 | dBWMR2 | 1 | vBWML6 | 1 | vBWML4 | 1 | dBWMR2 | 1 | dBWMR6 | 1 | mc1V | 1 | RMED |
| 2 | vm2PL | 2 | dBWMR4 | 2 | vBWMR1 | 2 | vBWML6 | 2 | dBWMR3 | 2 | ASEL | 2 | dBWMR1 | ||
| 3 | vm2PR | 3 | vBWML8 | 3 | vBWMR2 | 3 | vBWMR6 | 3 | dBWMR7 | 3 | AIZL | 3 | GLRL | ||
| 4 | vBWML10 | 4 | vBWMR6 | 4 | dBWMR9 | 4 | GLRDL | 4 | GLRR | ||||||
| 5 | vm1AL | 5 | dBWMR9 | 5 | vBWML8 | 5 | GLRDR | ||||||||
| 6 | vm1AR | 6 | hmc | 6 | vBWMR8 | 6 | hmc | ||||||||
| 7 | vm1PL | ||||||||||||||
| 8 | vm1PR |
Cell names corresponding to the indices in the heatmaps for wavenumber-dependent transmission map.
It is represented in the same way as Appendix 1—table 1.
| E = 3.000 | ka = 1.266 | E = 3.375 | ka = 1.227 | E = 3.475 | ka = 1.216 | E = 3.525 | ka = 1.211 | E = 3.625 | ka = 1.200 | E = 3.750 | ka = 1.186 | E = 3.875 | ka = 1.173 |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Index | Cell name | Index | Cell name | Index | Cell name | Index | Cell name | Index | Cell name | Index | Cell name | Index | Cell name |
| 0 | PLNR | 0 | ALA | 0 | PVDR | 0 | PVDL | 0 | ASKR | 0 | DVB | 0 | PHCR |
| 1 | PHAL | 1 | hmc | 1 | hmc | 1 | PVDR | 1 | PVQL | 1 | DD06 | 1 | PVR |
| 2 | PHCL | 2 | DVB | ||||||||||
| 3 | DA09 | 3 | PVT | ||||||||||
| 4 | PDB | 4 | AVL | ||||||||||
| 5 | PDA | ||||||||||||
| 6 | PDB | ||||||||||||
| 7 | VA12 | ||||||||||||
| 8 | VD12 |
List of cell-pairs with strong transmission except intra body-wall muscles pairs.
The first column represents the shortest distance along the network connecting the two cells of the cell-pair. The second and third columns represent the cell names of the cell-pair, and the color painted on the name box stands for the cell type (red: pharynx cells, orange: sensory neurons, yellow: inter neurons, green: motor neurons, light blue: body-wall muscles, purple: other end organs, magenta: sex-specific cells). The fourth, fifth, and sixth columns represent the transmission coefficient, the E value, and the wavenumber when the cell-pair shows the strongest transmission in the positive E value range, respectively.
| Distance | Cell names | Max{T(E)} | E(ka) | ka | Distance | Cell names | Max{T(E)} | E(ka) | ka | Distance | Cell names | Max{T(E)} | E(ka) | ka | |||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 17 | dBWMR24 | hmc | 0.912 | 0.125 | 1.558 | 3 | vBWMR10 | hmc | 0.791 | 0.325 | 1.538 | 2 | vm2AR | vm2PR | 0.892 | 1.700 | 1.400 |
| 17 | vBWMR24 | hmc | 0.765 | 0.325 | 1.538 | 3 | um1AL | vm2PR | 0.659 | 2.025 | 1.367 | 1 | ASEL | AFDL | 0.794 | 0.125 | 1.558 |
| 16 | vBWMR23 | hmc | 0.797 | 0.325 | 1.538 | 3 | um1AR | vm2PL | 0.659 | 2.025 | 1.367 | 1 | PVDL | PVDR | 0.858 | 0.750 | 1.496 |
| 15 | dBWMR22 | hmc | 0.805 | 0.125 | 1.558 | 3 | um1PL | vm2AR | 0.659 | 2.025 | 1.367 | 1 | PVDL | ALA | 0.704 | 0.850 | 1.486 |
| 15 | vBWMR22 | hmc | 0.603 | 0.325 | 1.538 | 3 | um1PR | vm2AL | 0.659 | 2.025 | 1.367 | 1 | PVDL | CANL | 0.776 | 0.750 | 1.496 |
| 14 | dBWMR21 | hmc | 0.946 | 0.125 | 1.558 | 2 | pm5VL | mc1V | 0.665 | 2.450 | 1.323 | 1 | PVDR | ALA | 0.728 | 0.750 | 1.496 |
| 12 | dBWMR19 | hmc | 0.892 | 0.125 | 1.558 | 2 | ASEL | AIZL | 0.839 | 2.450 | 1.323 | 1 | PVDR | CANR | 0.914 | 0.750 | 1.496 |
| 12 | vBWMR19 | hmc | 0.692 | 0.325 | 1.538 | 2 | ASKR | PVQL | 0.746 | 3.625 | 1.200 | 1 | PVDR | hmc | 0.627 | 3.475 | 1.216 |
| 11 | dBWMR18 | hmc | 0.731 | 0.125 | 1.558 | 2 | PLNR | PDB | 0.802 | 3.000 | 1.266 | 1 | PHBL | PHBR | 0.852 | 0.375 | 1.533 |
| 11 | vBWMR18 | hmc | 0.754 | 0.325 | 1.538 | 2 | PVDL | dBWML7 | 0.946 | 0.850 | 1.486 | 1 | CEPDL | OLQDL | 0.639 | 0.050 | 1.566 |
| 10 | dBWMR17 | hmc | 0.645 | 0.125 | 1.558 | 2 | PVDL | dBWML8 | 0.947 | 0.850 | 1.486 | 1 | IL1DR | IL1R | 0.642 | 0.225 | 1.548 |
| 10 | vBWMR17 | hmc | 0.789 | 0.325 | 1.538 | 2 | PVDL | CANR | 0.851 | 0.750 | 1.496 | 1 | IL1L | IL1VL | 0.599 | 0.225 | 1.548 |
| 9 | dBWMR16 | hmc | 0.936 | 0.125 | 1.558 | 2 | PVDR | dBWML7 | 0.737 | 0.850 | 1.486 | 1 | IL1R | IL1VR | 0.750 | 0.225 | 1.548 |
| 9 | vBWMR16 | hmc | 0.739 | 0.325 | 1.538 | 2 | PVDR | dBWML8 | 0.738 | 0.850 | 1.486 | 1 | IL1VL | IL1VR | 0.973 | 0.225 | 1.548 |
| 8 | vBWML15 | hmc | 0.593 | 0.275 | 1.543 | 2 | PVDR | CANL | 0.895 | 0.750 | 1.496 | 1 | IL1VR | URAVR | 0.556 | 0.225 | 1.548 |
| 7 | AVBR | dBWML12 | 0.601 | 1.000 | 1.471 | 2 | PHAL | DA09 | 0.546 | 3.000 | 1.266 | 1 | AIZL | AIZR | 0.534 | 0.375 | 1.533 |
| 7 | dBWMR14 | hmc | 0.867 | 0.125 | 1.558 | 2 | PHAL | PDB | 0.662 | 3.000 | 1.266 | 1 | ALA | CANR | 0.594 | 0.750 | 1.496 |
| 7 | vBWML14 | hmc | 0.726 | 0.225 | 1.548 | 2 | PHCL | PDB | 0.577 | 3.000 | 1.266 | 1 | PVPL | DVC | 0.568 | 0.475 | 1.523 |
| 7 | vBWML16 | hmc | 0.776 | 0.275 | 1.543 | 2 | IL1DR | IL1L | 0.781 | 0.225 | 1.548 | 1 | DVB | DD06 | 0.535 | 3.750 | 1.186 |
| 6 | dBWMR13 | hmc | 0.900 | 0.125 | 1.558 | 2 | IL1DR | IL1VR | 0.844 | 1.425 | 1.428 | 1 | RIGL | RIGR | 0.760 | 0.925 | 1.478 |
| 6 | vBWML13 | hmc | 0.832 | 0.225 | 1.548 | 2 | IL1L | IL1VR | 0.511 | 0.225 | 1.548 | 1 | AVKR | SMBDR | 0.895 | 0.200 | 1.551 |
| 6 | vBWML17 | hmc | 0.801 | 0.225 | 1.548 | 2 | IL1R | IL1VL | 0.824 | 0.225 | 1.548 | 1 | PVT | VD12 | 0.641 | 3.875 | 1.173 |
| 6 | vBWMR13 | hmc | 0.593 | 0.325 | 1.538 | 2 | IL1R | URAVR | 0.872 | 0.225 | 1.548 | 1 | RIVL | RIVR | 0.807 | 0.575 | 1.513 |
| 5 | CEPDL | IL1R | 0.716 | 0.050 | 1.566 | 2 | IL1VL | URAVR | 0.632 | 0.225 | 1.548 | 1 | VA12 | VD12 | 0.786 | 3.875 | 1.173 |
| 5 | AS11 | vBWMR4 | 0.574 | 1.000 | 1.471 | 2 | ALA | hmc | 0.560 | 3.375 | 1.227 | 1 | VB05 | VB06 | 0.950 | 0.050 | 1.566 |
| 5 | VD04 | vBWML5 | 0.617 | 0.075 | 1.563 | 2 | PVR | VD12 | 0.574 | 3.875 | 1.173 | 1 | dBWMR1 | GLRDL | 0.884 | 2.000 | 1.369 |
| 5 | vBWML12 | hmc | 0.785 | 0.275 | 1.543 | 2 | DVB | VA12 | 0.590 | 3.875 | 1.173 | 1 | dBWMR1 | GLRDR | 0.752 | 2.000 | 1.369 |
| 5 | vBWML18 | hmc | 0.503 | 0.225 | 1.548 | 2 | DVB | VD12 | 0.734 | 3.875 | 1.173 | 1 | vBWML5 | hmc | 0.790 | 0.325 | 1.538 |
| 5 | vBWMR12 | hmc | 0.743 | 0.325 | 1.538 | 2 | AVL | VA12 | 0.538 | 3.875 | 1.173 | 1 | vBWML7 | hmc | 0.662 | 0.325 | 1.538 |
| 4 | CEPDL | IL1DR | 0.757 | 0.050 | 1.566 | 2 | RMEL | RMED | 0.949 | 2.650 | 1.303 | 1 | vBWMR5 | hmc | 0.838 | 0.325 | 1.538 |
| 4 | OLQDL | IL1R | 0.716 | 0.050 | 1.566 | 2 | RMED | dBWMR1 | 0.501 | 2.650 | 1.303 | 1 | vBWMR7 | hmc | 0.573 | 0.325 | 1.538 |
| 4 | VD04 | vBWML7 | 0.535 | 0.075 | 1.563 | 2 | DA09 | PDB | 0.592 | 3.000 | 1.266 | 1 | dBWMR8 | hmc | 0.946 | 0.125 | 1.558 |
| 4 | dBWML4 | hmc | 0.721 | 0.000 | 1.571 | 2 | PDA | VA12 | 0.586 | 3.875 | 1.173 | 1 | vBWML8 | hmc | 0.774 | 0.225 | 1.548 |
| 4 | dBWMR11 | hmc | 0.913 | 0.125 | 1.558 | 2 | PDA | VD12 | 0.766 | 3.875 | 1.173 | 1 | CANL | exc_cell | 0.762 | 0.250 | 1.546 |
| 4 | vBWML11 | hmc | 0.705 | 0.275 | 1.543 | 2 | PDB | VD12 | 0.596 | 3.875 | 1.173 | 1 | CANR | exc_cell | 0.761 | 0.250 | 1.546 |
| 4 | vBWMR11 | hmc | 0.773 | 0.325 | 1.538 | 2 | VB05 | VB07 | 0.628 | 0.050 | 1.566 | 1 | mu_intL | mu_intR | 0.607 | 0.050 | 1.566 |
| 3 | PHCR | VD12 | 0.567 | 3.875 | 1.173 | 2 | vBWML4 | hmc | 0.803 | 0.325 | 1.538 | 1 | mu_intL | mu_anal | 0.984 | 0.050 | 1.566 |
| 3 | OLQDL | IL1DR | 0.757 | 0.050 | 1.566 | 2 | vBWML6 | hmc | 0.822 | 0.325 | 1.538 | 1 | mu_intL | mu_sph | 0.984 | 0.050 | 1.566 |
| 3 | IL1DR | URAVR | 0.708 | 0.225 | 1.548 | 2 | vBWMR4 | hmc | 0.793 | 0.325 | 1.538 | 1 | mu_intR | mu_anal | 0.625 | 0.050 | 1.566 |
| 3 | IL1L | IL1R | 0.859 | 0.225 | 1.548 | 2 | vBWMR6 | hmc | 0.992 | 1.800 | 1.390 | 1 | mu_intR | mu_sph | 0.625 | 0.050 | 1.566 |
| 3 | IL1L | URAVR | 0.871 | 0.225 | 1.548 | 2 | dBWMR9 | hmc | 0.807 | 0.125 | 1.558 | 1 | um1AL | um1PL | 0.573 | 2.025 | 1.367 |
| 3 | ALA | dBWML7 | 0.866 | 0.850 | 1.486 | 2 | vBWML8 | exc_gl | 0.715 | 0.800 | 1.491 | 1 | um1AR | um1PR | 0.573 | 2.025 | 1.367 |
| 3 | ALA | dBWML8 | 0.867 | 0.850 | 1.486 | 2 | vBWML9 | hmc | 0.707 | 0.225 | 1.548 | 1 | vm2AL | vm2AR | 0.927 | 1.700 | 1.400 |
| 3 | ALA | vBWML8 | 0.507 | 0.800 | 1.491 | 2 | GLRL | GLRR | 0.916 | 2.650 | 1.303 | 1 | vm1AL | vm1AR | 0.714 | 1.750 | 1.395 |
| 3 | AVFL | AS03 | 0.625 | 1.000 | 1.471 | 2 | CANL | CANR | 0.999 | 1.950 | 1.375 | 1 | vm1AL | vm1PL | 0.795 | 1.750 | 1.395 |
| 3 | dBWML5 | hmc | 0.806 | 0.000 | 1.571 | 2 | mu_anal | mu_sph | 0.984 | 0.050 | 1.566 | 1 | vm1AR | vm1PR | 0.795 | 1.750 | 1.395 |
| 3 | dBWMR1 | hmc | 0.869 | 2.000 | 1.369 | 2 | vm2AL | vm2PL | 0.892 | 1.700 | 1.400 | 1 | vm1PL | vm1PR | 0.714 | 1.750 | 1.395 |
| 3 | vBWML3 | hmc | 0.604 | 0.325 | 1.538 | 2 | vm2AL | vm2PR | 0.888 | 1.700 | 1.400 | 1 | vm2PL | vm2PR | 0.927 | 1.700 | 1.400 |
| 3 | vBWML10 | hmc | 0.797 | 0.225 | 1.548 | 2 | vm2AR | vm2PL | 0.888 | 1.700 | 1.400 | ||||||





