Observing one-divalent-metal-ion-dependent and histidine-promoted His-Me family I-PpoI nuclease catalysis in crystallo
Figures
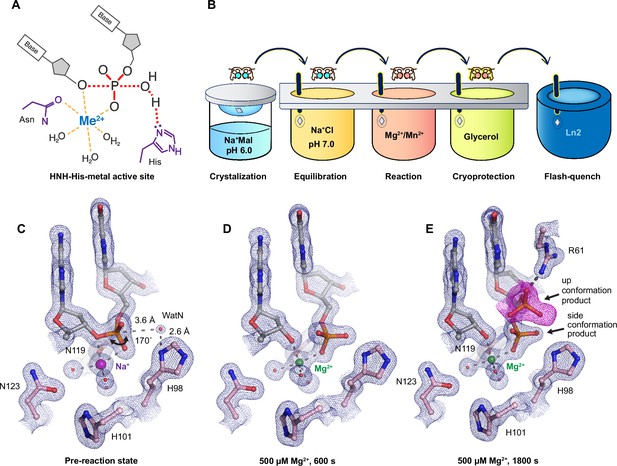
Observing histidine-metal (His-Me) family I-PpoI catalyze DNA hydrolysis in crystallo.
(A) Model of one-metal-ion-dependent and histidine-promoted His-Me enzyme catalysis and transition-state stabilization. (B) Metal ion soaking setup for in crystallo observation of DNA hydrolysis with I-PpoI. Structural intermediates of I-PpoI in crystallo DNA cleavage showcasing the pre-reaction state in (C) and product states in (D) and (E). The 2Fo − Fc map for Me2+, DNA, waters (red spheres), and catalytic residues (blue) was contoured at 2.0 σ (σ values represent root mean square (r.m.s). density values (later refered to as r.m.s.d.)). (E) The Fo − Fc omit map for the up conformation of the product (violet) was contoured at 3.0 σ.
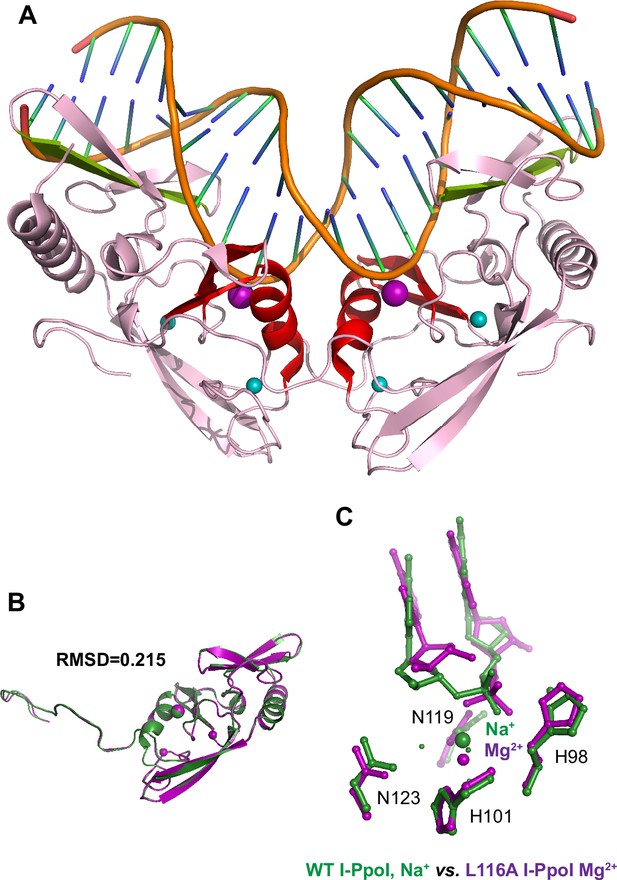
Overall structure and catalytic core of homing endonuclease I-PpoI.
(A) Homing endonuclease I-PpoI (PDB ID 1CZO) binding as a dimer to bend DNA at 55°. The overall endonuclease is colored in pink while the DNA is colored in orange. The catalytic core comprised of an alpha helix and two beta sheets are highlighted in red. The metal ion-binding site is depicted as a purple sphere while the Zn2+-binding sites are depicted as turquoise spheres. The beta sheets involved in DNA binding are depicted in light green. (B) Monomer of homing endonuclease I-PpoI (PDB ID 1CZO) (green) superimposed on top of the other I-PpoI monomer (purple) within the same unit cell, resulting in a RMSD of 0.215. (C) Structural comparison of the active site of WT I-PpoI (PDB ID 1CZ0) versus Leu116Ala I-PpoI (PDB ID 1EVW). The DNA fails to dock tightly toward the Mg2+ in the active site of Leu116Ala I-PpoI.
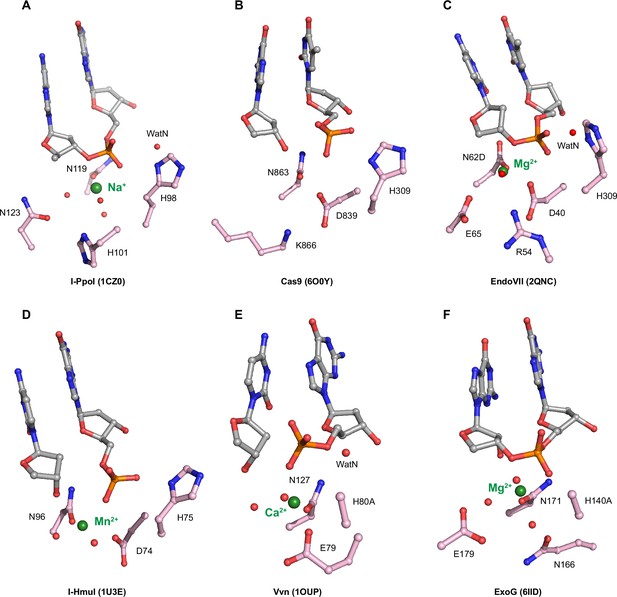
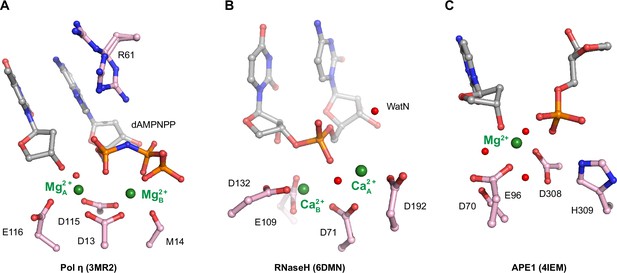
Active sites of histidine-metal (His-Me) superfamily nucleases.
Active sites of His-Me nucleases such as I-PpoI in (A), Cas9 in (B), EndoVII in (C), I-HmuI in (D), Vvn in (E), and ExoG in (F). Carbon atoms of residues within the active site are colored in pink. The metal ions are depicted by green spheres while waters are depicted by red spheres.
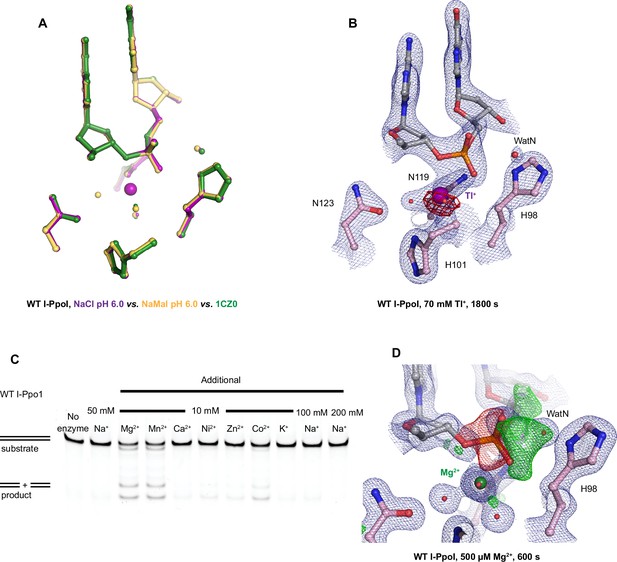
Establishing I-PpoI for in crystallo studies.
(A) Structural comparison of the active site after soaking in NaCl in purple, sodium malonate in yellow and PDB ID 1CZ0 in green. (B) Tl+ anomalous signal was detected at the I-PpoI metal ion-binding site after 1800s soaking in 70 mM Tl+. The 2Fo − Fc map for Me2+, DNA, waters (red spheres), and catalytic residues (blue) was contoured at 2.0 σ. The anomalous map for Tl+ was determined with X-ray at a wavelength of 0.9765 Å and contoured at 3.0 σ. (C) In solution metal ion assay of 10 mM additional metal ions on I-PpoI DNA hydrolysis. (D) Negative Fo − Fc peaks (red) were detected on the leaving group side of the scissile phosphate while positive Fo − Fc peaks (green) were detected on the nucleophile side after 600 s Mg2+ soaking. The 2Fo − Fc map for Me2+, DNA, waters (red spheres), and catalytic residues (blue) was contoured at 2.0 σ. The negative Fo − Fc map for the reactant phosphate (red) and the positive Fo − Fc map for the product phosphate (green) were contoured at 3.0 σ.
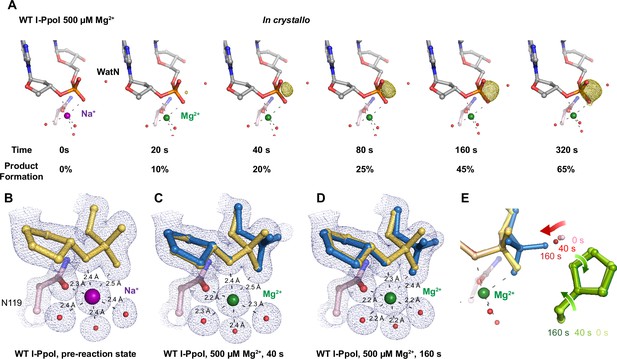
In crystallo DNA hydrolysis by I-PpoI.
(A) Structures of I-PpoI during in crystallo catalysis after 500 µM Mg2+ soaking for 0, 20, 40, 80, 160, and 320 s. The Fo − Fc omit map for the product phosphate (green) was contoured at 3.0 σ. I-PpoI complexes featuring metal ion coordination when bound with Na+ in (B) Mg2+ in the earlier time point of the reaction process in (C), and Mg2+ in the later time point of the reaction process in (D). The 2Fo − Fc map for Me2+, DNA, waters (red spheres), and catalytic residues (blue) was contoured at 2.0 σ. (E) Alignment of the WatN and rotation in His98 during I-PpoI reaction.
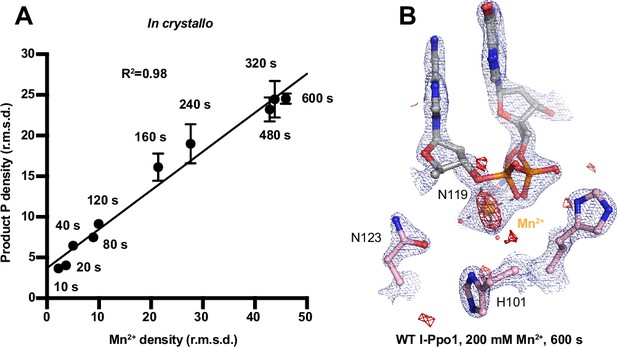
Detection of Me2+ binding during DNA hydrolysis in crystallo.
(A) Correlation (R2) between the newly formed phosphate and Mn2+ binding at pH 6. The points represent the mean of duplicate measurements for the electron density of the reaction product phosphate within two I-PpoI molecules in the asymmetric unit while the error bars represent the standard deviation. (B) Additional Mn2+-binding sites were not detected in the I-PpoI active site after 10 min soaking in 200 mM Mn2+. The 2Fo − Fc map for Me2+, DNA, waters (red spheres), and catalytic residues (blue) was contoured at 2.0 σ. The anomalous map for Mn2+ was collected at X-ray wavelength of 0.9765 Å and contoured at 3.0 σ.
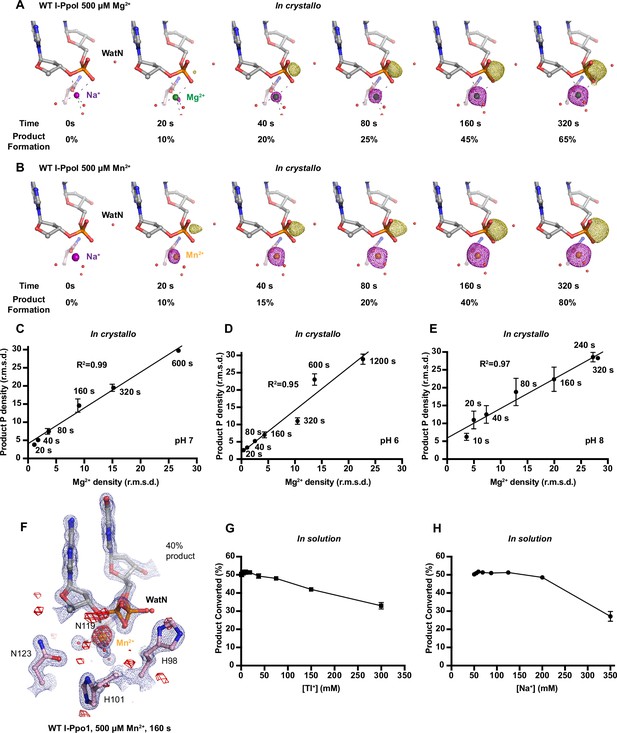
Additional metal ions are not required for DNA hydrolysis by I-PpoI.
(A) Structures of I-PpoI during in crystallo catalysis after 500 µM Mg2+ soaking for 0, 20, 40, 80, 160, and 320 s. The Fo − Fc omit maps for the product phosphate (green mesh) and Mg2+ (purple mesh) were contoured at 3.0 σ. (B) Structures of I-PpoI during in crystallo catalysis after 500 µM Mn2+ soaking for 0, 20, 40, 80, 160, and 320 s. The Fo − Fc omit maps for the product phosphate (green mesh) and Mn2+ (purple mesh) were contoured at 3.0 σ. Correlation (R2) between the newly formed phosphate and Mg2+ binding in crystallo at pH 7 in (C), pH 6 in (D), and pH 8 in (E). (C–E) The points represent the mean of duplicate measurements for the electron density of the reaction product phosphate within two I-PpoI molecules in the asymmetric unit while the error bars represent the standard deviation. (F) Mn2+ binding during DNA hydrolysis as revealed by anomalous signal of Mn2+ after 0.9786 Å X-ray diffraction. The 2Fo − Fc map for Me2+, DNA, waters (red spheres), and catalytic residues (blue) was contoured at 2.0 σ. The anomalous map for Mn2+ was contoured at 3.0 σ. (G) Tl+ concentration on in solution DNA cleavage. Precipitation was detected when Tl+ was at or greater than 150 mM. (H) Additional Na+ concentration on in solution DNA cleavage. (G, H) The error bars represent the standard deviation while the points represent the mean of triplicate measurements for cleaved DNA product.
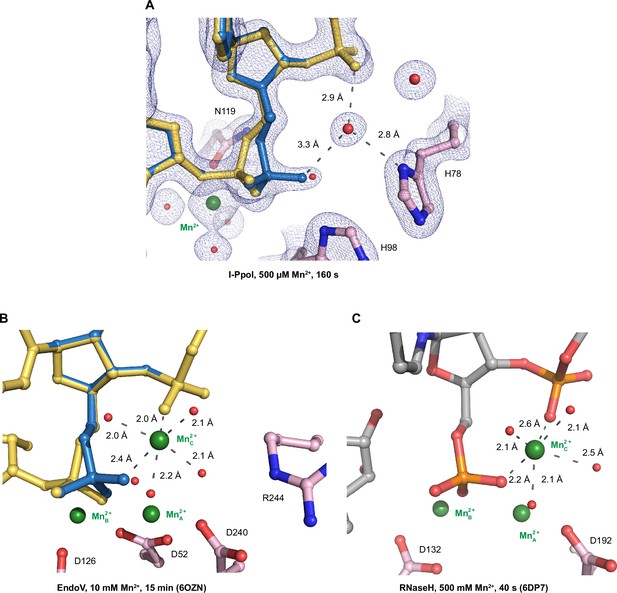
Environment of the transient Me2+ in nucleases.
Speculative ligand environment of the transient Me2+ in I-PpoI (A) in comparison to EndoV (B) and RNaseH (C). (A) The 2Fo − Fc map for Me2+, DNA, waters (red spheres), and catalytic residues (blue) was contoured at 2.0 σ. The DNA phosphate conformation within the active site of I-PpoI is looser in comparison to that in EndoV and RNaseH to bind an additional Me2+. Carbon atoms of residues within the active site are colored in pink. The Me2+ are depicted by green spheres while waters are depicted by red spheres. (A, B) The DNA reactant state is colored in yellow while the DNA product state is colored in blue.
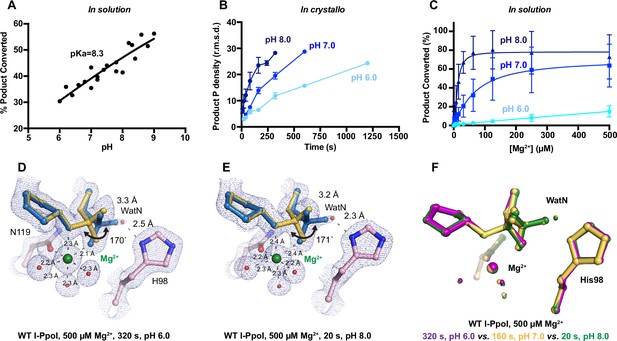
Effect of pH on I-PpoI DNA hydrolysis.
(A) DNA hydrolysis by WT I-PpoI with increasing pH in solution. (B) DNA hydrolysis by WT I-PpoI in crystallo at pH 6, 7, and 8. The points represent the mean of duplicate measurements for the electron density of the reaction product phosphate after a period of Mg2+ soaking within two I-PpoI molecules in the asymmetric unit. The error bars represent the standard deviation. (C) The effect of pH on metal ion dependence in solution. The points represent the mean of triplicate measurements for the percentage of cleaved DNA product while the error bars represent the standard deviation. (D) Structure of I-PpoI in crystallo DNA hydrolysis at pH 6 after 320 s of 500 µM Mg2+ soaking. (E) Structure of I-PpoI in crystallo DNA hydrolysis at pH 8 after 20 s of 500 µM Mg2+ soaking. (D, E) The 2Fo − Fc map for Me2+, DNA, waters (red spheres), and catalytic residues (blue) was contoured at 2.0 σ. (F) Structural comparison of the active site after 500 µM Mg2+ soaking for 320 s at pH 6 (violet), 160 s at pH 7 (yellow), and 20 s at pH 8 (green).
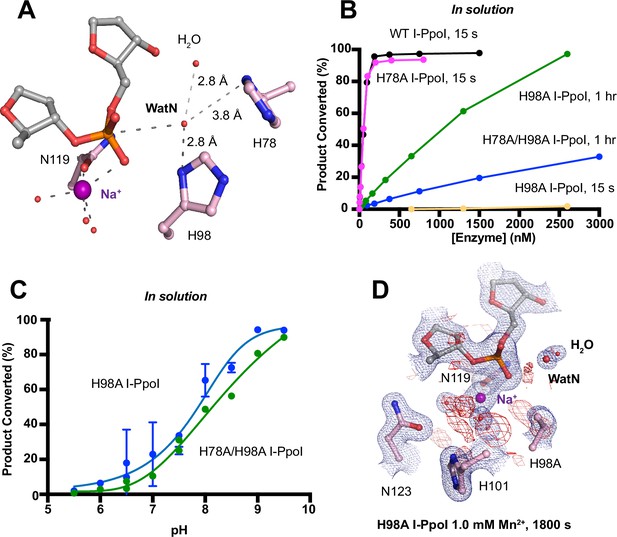
Nucleophilic water deprotonation during I-PpoI DNA hydrolysis.
(A) Active site environment surrounding WatN. His 78 exists near the WatN besides His98. (B) In solution DNA hydrolysis activity of various I-PpoI histidine mutants. The points represent the mean of triplicate measurements for the percentage of cleaved reaction product while the error bars (too small to see) represent the standard deviation. (C) DNA hydrolysis by H98A I-PpoI (blue) and H78A/H98A I-PpoI (green) at various pH in solution. (D) Structure of H98A I-PpoI active site after 1 mM Mn2+ soaking for 1800s. The anomalous map for Mn2+ was determined at X-ray wavelength of 0.9765 Å and contoured at 2.0 σ. The 2Fo − Fc map for Me2+, DNA, waters (red spheres), and catalytic residues (blue) was contoured at 2.0 σ. (B, C) The points represent the mean of triplicate measurements for the percentage of generated reaction product while the error bars represent the standard deviation.
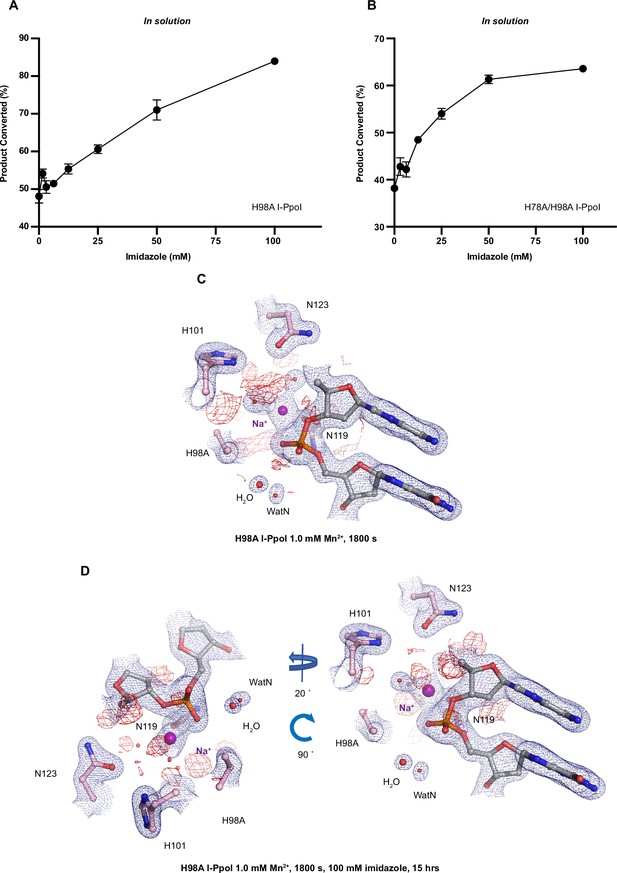
Histidine I-PpoI mutants and partial rescued cleavage activity by imidazole.
(A) In solution titration of imidazole on DNA cleavage activity by H98A I-PpoI. (B) In solution titration of imidazole on DNA cleavage activity by H78A/H98A I-PpoI. (A, B) The error bars represent the standard deviation while the points represent the mean of duplicate measurements for cleaved DNA product. (C) Structure of H98A I-PpoI active site after 1 mM Mn2+ soaking for 1800s. (D) Structure of H98A I-PpoI active site after 100 mM imidazole for 15 hr following 1 mM Mn2+ soaking for 1800s. The anomalous map for Mn2+ was contoured at 2.0 σ. (A, D) The 2Fo − Fc map for Me2+, DNA, waters (red spheres), and catalytic residues (blue) was contoured at 2.0 σ.
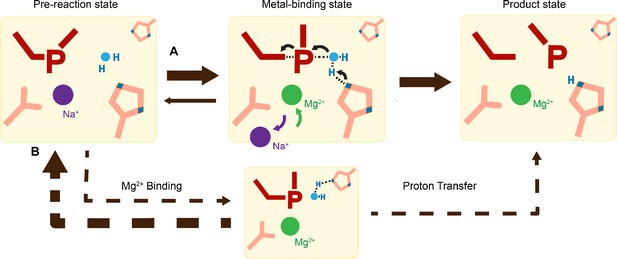
Catalytic model of histidine-metal (His-Me) nuclease DNA hydrolysis.
Proposed model of His-Me nuclease DNA hydrolysis in which Me2+ binding, proton transfer, and nucleophilic attack are concerted (solid arrow) in the presence of the primary proton acceptor in (A) versus unfavored (dashed arrows) in the absence of the primary proton acceptor in (B).
Additional files
-
Supplementary file 1
Crystal diffraction and refinement data.
- https://cdn.elifesciences.org/articles/99960/elife-99960-supp1-v1.docx
-
MDAR checklist
- https://cdn.elifesciences.org/articles/99960/elife-99960-mdarchecklist1-v1.docx