Peer review process
Revised: This Reviewed Preprint has been revised by the authors in response to the previous round of peer review; the eLife assessment and the public reviews have been updated where necessary by the editors and peer reviewers.
Read more about eLife’s peer review process.Editors
- Reviewing EditorKristin Tessmar-RaibleUniversity of Vienna, Vienna, Austria
- Senior EditorClaude DesplanNew York University, New York, United States of America
Reviewer #2 (Public Review):
In this revised manuscript Aguillon and collaborators convincingly demonstrating that CLK is required for free-running behavioral rhythms under constant conditions in the Cnidarian Nematostella. The results also convincingly show that CLK impacts rhythmic gene expression in this organism. This original work thus demonstrate that CLK was recruited very early during animal evolution in the circadian clock mechanism to optimize behavior and gene expression with the time-of-day. The manuscript could still benefit from some improvements so that it is more accessible for a wide readership.
Author response:
The following is the authors’ response to the original reviews.
eLife assessment
The study provides potentially fundamental insight into the function and evolution of daily rhythms. The authors investigate the function of the putative core circadian clock gene Clock in the cnidarian Nematostella vectensis. While it parts still incomplete, the evidence suggests that, in contrast to mice and fruit flies, Clock in this species is important for daily rhythms under constant conditions, but not under a rhythmic light/dark cycle, suggesting that the major role of the circadian oscillator in this species could be a stabilizing function under non-rhythmic environmental conditions.
Public Reviews:
Reviewer #1 (Public Review):
In this nice study, the authors set out to investigate the role of the canonical circadian gene Clock in the rhythmic biology of the basal metazoan Nematostella vectensis, a sea anemone, which might illuminate the evolution of the Clock gene functionality. To achieve their aims the team generated a Clock knockout mutant line (Clock-/- ) by CRISPR/Cas9 gene deletion and subsequent crossing. They then compared wild-type (WT) with Clock-/- animals for locomotor activity and transcriptomic changes over time in constant darkness (DD) and under light/dark cycles to establish these phenotypes under circadian control and those driven by light cycles. In addition, they used Hybridization Chain Reaction-In situ Hybridization (HCR-ISH) to demonstrate the spatial expression of Clock and a putative circadian clocl-controlled gene Myh7 in whole-mounted juvenile anemones.
The authors demonstrate that under LD both WT and Clock-/- animals were behaviourally rhythmic but under DD the mutants lost this rhythmicity, indicating that Clock is necessary for endogenous rhythms in activity. With altered LD regimes (LD6:6) they show also that Clock is light-dependent. RNAseq comparisons of rhythmic gene expression in WT and Clock-/- animals suggest that clock KO has a profound effect on the rhythmic genome, with very little overlap in rhythmic transcripts between the two phenotypes; of the rhythmic genes in both LD and DD in WT animals (220- termed clock-controlled genes, CCGS) 85% were not rhythmic in Clock-/- animals in either light condition. In silico gene ontology (GO) analysis of CCGS reflected process associated with circadian control. Correspondingly, those genes rhythmic in KO animals under DD (here termed neoCCGs) were not rhythmic in WT, lacked upstream E-box motifs associated with circadian regulation, and did not display any GO enrichment terms. 'Core' circadian genes (as identified in previous literature) in WT and Clock-/- animals were only rhythmic under entrainment (LD) conditions whilst Clock-/- displayed altered expression profiles under LD compared to WT. Comparing CCGs with previous studies of cycling genes in Nematostellar, the authors selected a gene from 16 rhythmic transcripts. One of these, Myh7 was detectable by both RNAseq and HCR-ISH and considered a marker of the circadian clock by the authors.
The authors claim that the study reveals insights into the evolutionary origin of circadian timing; Clock is conserved across distant groups of organisms, having a function as a positive regulator of the transcriptional translational feedback loop at the heart of daily timing, but is not a central element of the core feedback loop circadian system in this basal species. Their behavioural and transcriptomic data largely support the claims that Clock is necessary for endogenous daily activity but that the putative molecular circadian system is not self-sustained under constant darkness (this was known already for WT animals)- rather it is responsive to light cycles with altered dynamics in Clock-/- specimens in some core genes under LD. In the main, I think the authors achieved their aims and the manuscript is a solid piece of important work. The Clock-/- animal is a useful resource for examining time-keeping in a basal metazoan.
The work described builds on other transcriptomic-based works on cnidaria, including Nematostellar, and does probe into the molecular underpinnings with a loss-of-function in a gene known to be core in other circadian systems. The field of chronobiology will benefit from the evolutionary aspect of this work and the fact that it highlights the necessity to study a range of non-model species to get a fuller picture of timing systems to better appreciate the development and diversity of clocks.
Strengths:
The generation of a line of Clock mutant Nematostellar is a very useful tool for the chronobiological community and coupled with a growing suite of tools in this species will be an asset. The experiments seem mostly well conceived and executed (NB see 'weaknesses'). The problem tackled is an interesting one and should be an important contribution to the field.
Weaknesses:
I think the claims about shedding light on the evolutionary origin of circadian time maintenance are a little bold. I agree that the data do point to an alternative role for Clock in this animal in light responsiveness, but this doesn't illuminate the evolution of time-keeping more broadly in my view. In addition, these are transcriptomic data and so should be caveated- they only demonstrate the expression of genes and not physiology beyond that. The time-course analysis is weakened by its low resolution, particularly for the RAIN algorithm when 4-hour intervals constrain the analysis. I accept that only 24h rhythms were selected in the analysis from this but, it might be that detail was lost - I think a preferred option would be 2 or 3-hour resolution or 2 full 24h cycles of analysis.
The authors discount the possibility of the observed 12h rhythmicity in Clock-/- animals by exposing them to LD6:6 cycles before free-running them in DD. I suggest that LD cycles are not a particularly robust way to entrain tidal animals as far as we know. Recent papers show inundation/mechanical agitation are more reliable cues (Kwiatkowski ER, et al. Curr Biol. 2023, 2;33(10):1867-1882.e5. doi: 10.1016/j.cub.2023.03.015; Zhang L., et al Curr Biol. 2013, 23;19, 1863-1873 doi.org/10.1016/j.cub.2013.08.038.) and might be more effective in revealing endogenous 12h rhythms in the absence of 24h cues.
Response: We removed the suggestion that we used 6:6h LD to perform tidal entrainment. We generated this ultradian light condition to address the 24h rhythmicity observed in the NvClk1-/- in 12:12h LD.
Reviewer #2 (Public Review):
This manuscript addresses an important question: what is the role of the gene Clock in the control of circadian rhythms in a very primitive group of animals: Cnidaria. Clock has been found to be essential for circadian rhythms in several animals, but its function outside of Bilaterian animals is unknown. The authors successfully generated a severe loss-of-function mutant in Nematostella. This is an important achievement that should help in understanding the early evolution of circadian clocks. Unfortunately, this study currently suffers from several important weaknesses. In particular, the authors do not present their work in a clear fashion, neither for a general audience nor for more expert readers, and there is a lack of attention to detail. There are also important methodological issues that weaken the study, and I have questions about the robustness of the data and their analysis. I am hoping that the authors will be able to address my concerns, as this work should prove important for the chronobiology field and beyond. I have highlighted below the most important issues, but the manuscript needs editing throughout to be accessible to a broad audience, and referencing could be improved.
Major issues:
(1) Why do the authors make the claim in the abstract that CLOCK function is conserved with other animals when their data suggest that it is not essential for circadian rhythms? dCLK is strictly required in Drosophila for circadian rhythms. In mammals, there are two paralogs, CLOCK and NPAS2, but without them, there are no circadian rhythms either. Note also that the recent claim of BMAL1-independent rhythms in mammals by Ray et al., quoted in the discussion to support the idea that rhythms can be observed in the absence of the positive elements of the circadian core clock, had to be corrected substantially, and its main conclusions have been disputed by both Abruzzi et al. and Ness-Cohn et al. This should be mentioned.
Response: According to our Behavioral and Transcriptomic data, CLOCK function is conserved in constant light condition. In LD context, the rhythmicity is maintained probably by the light-response pathway in Nematostella. We modified our rhythmic transcriptomic analysis and considered the context of the contested results by Ray et al., and discussed it in the revised manuscript.
(2) The discussion of CIPC on line 222 is hard to follow as well. How does mRNA rhythm inform the function of CIPC, and why would it function as a "dampening factor"? Given that it is "the only core clock member included in the Clock-dependent CCGs," (220) more discussion seems warranted. Discussing work done on this protein in mammals and flies might provide more insight.
Response: The initial sentence was unclear. Furthermore, since we restricted our rhythmic analysis to genes only found rhythmic with a p<0.01 with RAIN combined with JTK, NvCipc was no longer defined as rhythmic in free running.
(3) The behavioral arrhythmicity seen with their Clock mutation is really interesting. However, what is shown is only an averaged behavior trace and a single periodogram for the entire population. This leaves open the possibility that individual animals are poorly synchronized with each other, rather than arrhythmic. I also note that in DD there seem to be some residual rhythms, though they do not reach significance. Thus, it is also possible that at least some individual animals retain weak rhythms. The authors should analyze behavioral rhythms in individual animals to determine whether behavioral rhythmicity is really lost. This is important for the solidity of their main conclusions.
Response: Fig. 1 has been modified. We have separated the data for WT and NvClk1-/- animals to provide clarity on the average behavior pattern for each genotype. While the LSP analysis on the population average informs us about the synchronization of the population, it is true that it does not provide insight into individual rhythmicity. To address this, we analyzed individuals in all conditions using the Discorhythm website (Carlucci et al., 2019).
In the revised figure, we have included a comparison plot of the acrophase of 24-hour rhythmic animals between genotypes using Cosinor analysis, which is most suitable for acrophase detection. This plot indicates the number of animals detected as significantly rhythmic, providing direct visual input to the reader regarding individual rhythmicity. Additionally, we have added Table 1, which contains the Cosinor period analysis (24 and 12 hours) of individuals for all genotypes and conditions, further enhancing the clarity of our findings.
(4) There is no mention in the results section of the behavior of heterozygotes. Based on supplement figure 2A, there is a clear reduction in amplitude in the heterozygous animals. Perhaps this might be because there is only half a dose of Clock, but perhaps this could be because of a dominant-negative activity of the truncated protein. There is no direct functional evidence to support the claim that the mutant allele is nonfunctional, so it is important to discuss carefully studies in other species that would support this claim, and the heterozygous behavior since it raises the possibility that the mutant allele acts as a dominant negative.
Response: Extended Data Fig.1 modified. We show NvClk1+/- normalized locomotion over time in DD of the population, comparison of individual normalized behavior amplitude, LSP of the average population and individual acrophase of only rhythmic 24h individuals. Indeed, we cannot discriminate Dominant-negative from non-functional allele.
(5) I do not understand what the bar graphs in Figure 2E and 3B represent - what does the y-axis label refer to?
Response: Not relevant to the revised manuscript.
(6a) I note that RAIN was used, with a p<0.05 cut-off. I believe RAIN is quite generous in calling genes rhythmic, and the p-value cut-off is also quite high. What happens if the stringency is increased, for example with a p<0.01.
Response: We acknowledge your concern regarding the stringency of our statistical analysis. To address this, we opted to combine both RAIN and JTK methods and applied a more stringent p-value cut-off of p<0.01.
(6b) It would be worth choosing a few genes called rhythmic in different conditions (mutant or wild-type. LD or DD), and using qPCR to validate the RNAseq results. For example, in Figure 3D, Myh7 RNAseq data are shown, and they do not look convincing. I am surprised this would be called a circadian rhythm. In wild-type, the curve seems arrhythmic to me, with three peaks, and a rather large difference between the first and second ZT0 time point. In the Clock mutants, rhythms seem to have a 12hr period, so they should not be called rhythmic according to the material and methods, which says that only ca 24hr period mRNA rhythms were considered rhythmic. Also, the result section does not say anything about Myh7 rhythms. What do they tell us? Why were they presented at all?
Response: Regarding the suggestion for independent verification of our RNAseq results, we agree that such validation would enhance the robustness of our findings. To address this, we chose to overlap our identified rhythmic genes under WT LD conditions with those from another transcriptomic study that shared similarities in experimental design. Notably, the majority of overlapping rhythmic genes between the studies are candidate pacemaker genes. We believe that this replication of biologically significant rhythmic genes strengthens the validity and reliability of our results (see Extended Data Fig. 2).
Furthermore, we have decided to remove the NvMhc-st (mistakenly named Myh7, only rhythmic in WT DD in the new analysis) as it does not contribute substantively to the revised version of the manuscript.
(7) The authors should explain better why only the genes that are both rhythmic in LD and DD are considered to be clock-controlled genes (CCGs). In theory, any gene rhythmic in DD could be a CCG. However, Leach and Reitzel actually found that most genes in DD1 do not cycle the next day (DD2)? This suggests that most "rhythmic" genes might show a transient change in expression due to prolonged obscurity and/or the stress induced by the absence of a light-dark cycle, rather than being clock controlled. Is this why the authors saw genes rhythmic under both LD and DD as actual CCGs? I would suggest verifying that in DD the phase of the oscillation for each CCG is similar to that in LD. If a gene is just responding to obscurity, it might show an elevated expression at the end of the dark period of LD, and then a high level in the first hours of DD. Such an expression pattern would be very unlikely to be controlled by the circadian clock.
Response: As we modified our transcriptomic analysis, we do no longer analyze LD+DD rhythmic genes, but any genes rhythmic (RAIN and JTK p<0.01) in each condition. As such we end up with four list of genes corresponding to each experimental conditions.
(8) Since there are still rhythms in LD in Clock mutants, I wonder whether there is a paralog that could be taking Clock's place, similar to NPAS2 in mammals.
Response: see response to (1) > The only NPAS2 orthologous identified in Nematostella NPAS3 showed marginally significance (p=0.013) with RAIN in LD WT suggesting a regulation similar to the candidate pacemaker genes. As such we included within our candidate pacemaker genes list.
(9) I do not follow the point the authors try to make in lines 268-272. The absence of anticipatory behavior in Drosophila Clk mutants results from disruption of the circadian molecular clock, due to the loss of Clk's circadian function. Which light-dependent function of Clock are the authors referring to, then? Also, following this, it should be kept in mind that clock mutant mice have a weakened oscillator. The effect on entrainment is secondary to the weakening of the oscillator, rather than a direct effect on the light input pathway (weaker oscillators have increased response to environmental inputs). The authors thus need to more clearly explain why they think there is a conservation of circadian and photic clock function.
Response: Following the changes in our statistical analysis we reframed the discussion and address directly the circadian and the photic clock function (we call it light-response pathway in the manuscript)
Recommendations for the authors:
We suggest the following improvements:
(1) Please undertake a serious effort to make this work more accessible to non-marine chronobiologists. This includes better explanations, and schemes of the animal when images of staining are shown (e.g. Fig.1b) which include the labeling of relevant morphological structures mentioned in the text (like "tentacle endodermis and mesenteries" (line 132)). Similar issues for mentioned life cycle stages like "late planula stage" (line 133), "bisected physa" (line 149).
Response: Fig. 1b, we outlined the animal shaped and added 2 arrows to locate the tentacle endodermis and mesenteries. We replaced the term late planula stage, by larvae. And we rephrased bisected physa by tissue sampling.
Please attend to details. This includes:
- Wrong referrals to figures (currently line 151 refers to EDF2- but should be EDF 1 instead, there is a Fig.3f mentioned in the text, but there is no such Fig.).
Response: Fixed
- Mentioning of ZTs when the HCR stainings were performed.
Response: Fixed
- Fig.1 a shows a rather incomplete and thus potentially confusing phylogenetic tree. Vertebrates have at least two Clk orthologs (NPAS2 and CLK), please include both, use an outgroup, and rout the tree.
Response: Identifying NPAS2 and CLK orthologous in all species added more confusion into the conclusion. However, we followed the suggestion of adding an outgroup using a CLK orthologous sequence identified in the sponge Amphimedon queenslandica and rout the tree. Thank for the suggestion.
- What do the y-axis labels in Figure 2E and 3B refer to exactly? Y-axis label annotations in Fig.3a,d are entirely missing- what do the numbers refer to?
Response: not relevant in the revised manuscript
- Fig.2D- is the Go term enrichment referring to LD or DD?
Response: to DD. We made it cleared on the figure 5.
- Wording: "Clock regulates genetic pathways." What is meant by "genetic pathways"? There are no "non-genetic pathways". Could one simply say: "Clock regulates a variety of transcripts".
Response: We modified our threshold to use only p.adj<0.01, which reduced the GO term numbers. We removed “genetic pathways” and now address the specific pathways: cell-cycle and neuronal.
The use of the term "epistatic" is confusing (line 219), i.e. that light is epistatic to Clock. In genetics, epistasis is defined as the effect of gene interactions on phenotypes. To a geneticist, this implies that there is a second gene impacting on the phenotype of the Clock mutants. Please re-word.
Response: “light is epistatic on Clock” has been re-phrased.
The provided Supplementary tables are not well annotated. Several of them need guess-work about what is shown. For instance, for Supplementary Table 1, the Ns are unclear, which in total can go up to almost 200 per condition-genotype, but only about 30 animals for each were tested. Thus, where do the high totals in the LSP table come from? What do the numbers of each periodicity mean? Initially one might assume it was the number of animals that showed a periodogram peak at a given periodicity, but it seems that cannot be. Maybe it counted any period bin over statistical significance? Please clarify with better descriptions and labels.
Response: Supplementary tables are now clearly annotated on their first Tabs. About Fig.1, we already addressed this point in the public review.
Albeit not essential, it would be more reader-friendly to also add a summary table with average period and SD, power and SD, and percentage rhythmicity to the main figure.
Response: Table 1 is added: it contains individual count of rhythmic animals (24h and 12h) with Cosinor. However, using Discorhythm we had to ask for a specific Period. Thus, we can only provide animal count significant for a given period value. And not an estimation of their own period.
(2) Some of the terminology is quite confusing, in particular the double meaning of the word "clock" (i.e the pacemaker and the transcription factor). This is not a specific problem to this manuscript, but it would be helpful for the readability to try to improve this.
Could the gene/transcript/protein be spelled: clk and Clk?
Alternatively, for clarity- how about talking about "core pacemaker genes," "CLOCK-dependent rhythmic genes" and "CLOCK-independent rhythmic genes"?
Response:
Clock/CLOCK > NvClk / NvCLK and the mutant is NvClk1-/-
Core clock genes > candidate pacemaker genes.
CLOCK-dependent CCG > this notion no longer exists in the revised manuscript.
CLOCK-independent CCG > this notion no longer exists in the revised manuscript.
(3) The dismissal of the 12h rhythmicity in Clock-/- animals is not really convincing and should be reconsidered. LD6:6 cycles (before free-running animals in DD) is likely a not particularly robust way to entrain tidal animals. Recent papers show inundation/mechanical agitation are more reliable cues (Kwiatkowski ER, et al. Curr Biol. 2023, 2;33(10):1867-1882.e5. doi: 10.1016/j.cub.2023.03.015; Zhang L., et al Curr Biol. 2013, 23;19, 1863-1873 doi.org/10.1016/j.cub.2013.08.038.) and might be more effective in revealing endogenous 12h rhythms in the absence of 24h cues.
Response: We removed the proposition of using 6:6hLD as Tidal entrainment. Instead, the LD 6:6 experiment reveals the direct light-dependency of the NvClk1-/- mutant.
(4) There are significant questions raised on the validity of BMAL1-independent rhythms in mammals as suggested by the Ray et al study. See DOI: 10.1126/science.abe9230 and DOI: 10.1126/science.abf0922
These technical comments should also be taken into account and the discussion adjusted accordingly to better reflect the ongoing discussions in the chronobiology field.
Response: We modified our rhythmic analysis. As we cannot use BHQ or adjusted p-value which resulted in very genes, we defined 24h-rhythmic genes if p<0.01 with two different algorithms (RAIN and JTK). We propose this compromise to reduce the risk of false-positive. Furthermore, we discussed our methodology in the light of the significant questions raised by these papers you cited. We thank the reviewer for this important point.
(5) The HCR stainings for clk are not very convincing. Normally, HCR should have more dots. In principle, the logic of HCR is such that it detects individual mRNA molecules in the cell. Thus, having only one strong dot/cell like in Fig.1b doesn't make much sense.
Response: We were the first surprised by this single dot signal. We are experienced users of HCRv.3 across different species. We decided to remove the close-up (for further investigations) but to keep the full animal signal. According to our approach it is a convincing signal. However, the doty nature of the signal itself it is not easy to make it highly visible at full scale animal on the picture. We did our best to show the mRNA signal visible without altering the pattern.
Furthermore, the controls for the HCR in situ hybridization are unclear. In the methods, there are two Clock probes described (B3 & B5) and two control probes (B1 & B3), however, in the negative control image, a combination of one Clock (B1) and one control (B3) probes is used and is unclear what "redundant detection" means in the legend of figure S2.
Response: Considering the nature of the signal (single of few dots), we decided to use two probes with 2 different fluorophores. A noise is by nature random. Our hypothesis was: only overlapping fluorescent dots are true signal of NvClk mRNA.
For Control probes we used two zebrafish probes labelling hypothalamic peptides.
Based on the experience with non-Drosophila, non-mouse animal model systems the reviewers assume that non-sense mediated mRNA decay (NMD) is not strongly initiated upon Crispr-induced premature STOP-codons. If this assumption is correct it would be worth to mention it. Alternatively, it would be worth testing if Nematostella induces NMD, as this would be a great control for the HCR and the mutation itself. At which ZT was the HCR done?
Response: We performed the HCR at ZT10 when NvClk is described to be at peak. It is now indicated in the Fig. 1b. The RNAseq detected a higher quantity of NvClk1 mRNA in the NvClk1-/- (see Fig. 4a). mRNA quantity regulation involves transcription, stabilization, and degradation. At this stage, we cannot identify which specific step is affected.
For Fig.1c- please provide the binding site and sequence in the figure, simply include EDF 1 in the main figure.
Response: We generated a clear indication in the new Fig.1c and EDF. 1b about the protein domains, the CRISPR binding site and the consequences on the DNA and AA sequences.
(6) Please provide the individual trace data for the behavioral analyses either as supplementary files or as a link to an openly accessible database like DRYAD (see also comment 7 in the public review of reviewer 2). Maybe this is what is shown in Supplementary Table 1, but it is really not clear what is actually shown.
Response: Fig.1 is updated. Table 1 is added. Supplementary Table 1 contains individual normalized locomotor data of each polyps for each genotypes and light conditions. Supplementary Table 2 contains the cosinor individual rhythmic behavior analysis based on the Supplementary Table 1.
(7) It is not really clear if the mutation is a true loss-of-function or could also be dominant negative. While this is raised in the discussion, it should be more carefully considered. The reason why a dominant negative would be unlikely is unclear. More specifically also see comment 8) in the public review of reviewer 2.
Response: Indeed, the results cannot tell us if it is a true loss of function, a dominant negative or non-functional allele. We addressed it in the first part of the discussion.
(8) The pretty small overlap of rhythmic transcripts in LD and DD could reflect the true biology of a more core clock driven-process under constant conditions and a more light-driven process under LD. But still- wouldn't one expect that similar processes should be rhythmic? If not, why not?
It would certainly add strength to the data if for one or two transcripts these results were independently verified by qPCR from an independent sampling. This could even be done for just two time points with the most extreme differences.
Response: We appreciate the reviewer's comments and concerns regarding the overlap of rhythmic transcripts in different conditions. In response to the reviewer's query, we revised our interpretation of the transcriptomic data, acknowledging the limited overlap between light and genotype conditions in our study. This prompted us to reconsider the underlying biological processes driving rhythmic gene expression under constant conditions versus light-dark cycles.
Regarding the suggestion for independent verification of our RNAseq results, we agree that such validation would enhance the robustness of our findings. To address this, we chose to overlap our identified rhythmic genes under WT LD conditions with those from another transcriptomic study that shared similarities in experimental design. Notably, the majority of overlapping rhythmic genes between the studies are candidate pacemaker genes. We believe that this replication of biologically significant rhythmic genes strengthens the validity and reliability of our results (see Extended Data Fig. 2).
(9) Expression of myh7 : Checking for co-expression should be pretty straightforward by HCR. This is what this type of staining technique is really good for. Please do clk and myh7 co-staining if you want to claim co-expression. Otherwise don't make such a claim.
Response: We agree that checking for co-expression should be straightforward by HCR. However, due to time constraints during the revision period, we are unable to conduct the double in-situ experiment. Additionally, upon careful consideration, we recognize that including myhc-st (mistakenly named myh7) staining and co-expression analysis would not significantly contribute to the main conclusions of our study. Therefore, we have decided to remove this analysis from the revised manuscript.
(10) Missing methodological details:
- The false discovery rate for each analysis should be included (see Hughes et al.,: "Guidelines for Genome-Scale Analysis of Biological Rhythms," 2017).
Response: THE FDR is indicated for each gene in supplementary table 3
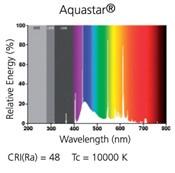
- Fig.1f- continuous light- please provide a spectrum (If there is no good spectrophotometer available, please provide at least manufacturer information.
Response: Unfortunately, we don’t have a good spectrophotometer available during the time of the revision. We added to the method the reference of the lamp. We found the light spectrum provided by the supplier. However, we did not add it to the revised manuscript.
Author response image 1.
Spectrum of the Aquastar t8
Also, it would be easier for the reader, if the measurements of light intensity are provided in photons, because this is what the light receptors ultimately measure.
Response: Modified.
- Fig.2E- please add the consensus sequence used for circadian E-box vs. E-box to the figure.
Response: In the revised manuscript Fig.4c, we show which E-box motifs we extracted for our promoter analysis. We as well changed our analysis and did no longer use HOMER, but we directly extracted promoter sequences and looked for canonical Ebox CANNTG and Circadian Ebox CACGTG and generate a Circadian Ebox enrichment output per gene promoter.
(11) There has been some discussion about the evolutionary statement as stated by the authors. It appears that depending on the background of the reader, this can be misunderstood. We thus suggest to more clearly point out where the author thinks there is evolutionary conservation (a function for clk in the circadian oscillator under constant light or dark conditions) versus where there is no apparent evolutionary conservation (the situation under light-dark conditions).
Response: In the revised manuscript we proposed a conserved function of NvCLK in constant darkness, and a light-response pathway compensating in LD conditions in the mutant.
Please also consider the major comments 8 and 9 of the common review from reviewer 2.
Reviewer #1 (Recommendations For The Authors):
The hybridization chain-reaction ISH is OK but, I'm not sure I understand the control condition-this should be clarified. I would also welcome the use of Clock-/- animals in HCR as another, more direct level of control. In addition, the authors state that the Myh7 probes hybridise in anatomical regions resembling those for Clock (Fig 3e). It would be better to duplex these two probe sets with different fluors for a better representation of the relative spatial distributions of each transcript.
Response: We agree that checking for co-expression should be straightforward by HCR. However, due to time constraints during the revision period, we are unable to conduct the double in-situ experiment. Additionally, upon careful consideration, we recognize that including myhc-st (mistakenly named myh7) staining and co-expression analysis would not significantly contribute to the main conclusions of our study. Therefore, we have decided to remove this analysis from the revised manuscript.
We clarified in the methods the control probes design.
Minor points:
Figure legends do not all convey sufficient detail. For instance, Figure 1c needs a better explanation. Figure 3e- are these images both WT? Fig 3f doesn't exist and other figure text references do not align with figures and need an overhaul.
Response: All errors have been fixed.
Reviewer #2 (Recommendations For The Authors):
Major issues:
(1) The authors need to introduce their model system better for a broad audience. What are the tissues/cells that express Clock at a higher level? What is their function, does this provide a potential explanation for their specific Clock expression, and how CLOCK might regulate behavior? Terms such as "tentacle endodermis and mesenteries" (line 132), "late planula stage" (line 133), "bisected physa" (line 149) would need some explanation.
Response: We modified term such as planula to larvae, and bisected physa to tissue samples.
- Some of the terminology used is quite confusing, because of the double-meaning of the word "clock" (i.e the pacemaker and the transcription factor). The authors use terms such as "clock-controlled genes", "core clock genes", "CLOCK-dependent clock-controlled genes", "neo-clock-controlled genes". Is there any way to help the reader? Here are several suggestions: "core pacemaker genes," "CLOCK-dependent rhythmic genes" and "CLOCK-independent rhythmic genes".
Response: all the terminology has been clarified, see previous comments
- Also in the abstract, there is mention of "hierarchal light- and Clock-signaling" (52-3) - is this related to the statement on line 219 that light is epistatic to Clock? I do not quite understand what epistatic would mean here. Who is upstream of whom? LD modifies rhythmicity in Clock mutant animals, but Clock mutations also impact rhythmicity in LD. Also, as epistasis is defined as the effect of gene interactions on phenotypes - what is the secondary gene impacting the phenotype of the Clock mutants? I am not sure the term epistatic is appropriate in the present context.
Response: Indeed, Epistatic is a genetic term which might be unclear in this context. We removed it.
- The control for the in situ hybridization is unclear. In the methods, there are two Clock probes described (B3 & B5) and two control probes (B1 & B3), however, in the negative control image, a combination of one Clock (B1) and one control (B3) probe is used, I am not sure what "redundant detection" means in the legend of figure S2. Also, the sequences of each Clock probe should be provided. It might be worth testing the Clock mutant the authors generated. Clock mRNA could be reduced due to non-sense, mediated RNA decay, since the mutation causes a premature stop codon. This would be a great additional control for the in situ hybridization. Even better would be if, by chance, the probes target the mutated sequence. The signal should then be completely lost.
Response: HCR is a tilling probe. Which means the target transcript is covered by dozens of successive DNA sequence “primer-like” which allow the HCRv.3 technology. We cannot design a mutant probe specific with this technology.
(5) I have concerns with rhythmic-expression calls, particularly as there is so little overlap between LD and DD, and that a completely different set of rhythmic genes is observed in Clock mutant and wild-type animals. I am not an expert in whole-genome expression studies, so I hope one of my colleague reviewers can weigh in.
When describing rhythmicity analysis in the Methods, it states that Benjamini-Hochberg corrections were applied to account for multiple comparisons. However, the false discovery rate for each analysis should be included (see Hughes et al.,: "Guidelines for Genome-Scale Analysis of Biological Rhythms," 2017).
Response: As explained before we cannot used Benjamini-Hochberg corrections as only few genes (mostly oscillator gene pass the threshold). As such we combined two different algorithms (RAIN and JTK) with a p<0.01 to detect confidently rhythmic genes while reducing the risk of false-positives.
Minor issues:
(1) Environmental inputs are not "circadian", as written in the title.
Response: Title modified
(2) In the abstract, the description of the Clock mutant behavioral phenotypes is hard to follow, with no mention of whether or not Clock mutant animals are behaviorally rhythmic or arrhythmic in constant conditions.
Response: corrected
(3) Abstract: A 6/6 h LD cycle is not a compressed tidal cycle as written in the abstract. Light is not an input to tidal rhythms.
Response: corrected
(4) Line 101: timeout is not a core clock gene in animals.
Response: we removed it from the candidate pacemaker genes.
(5) What is the evidence for the role of PAR-Zip proteins in the Nematostella clock? The reference provided does not mention those.
Response: There is no functional data in Nematostella yet to support their role within the pacemaker. However based on their rhythmicity in LD and protein conservation, we included them within the candidate pacemaker genes list. The refences have been corrected.
(6) Line 125. should refer to Fig 1C when describing the Clock protein.
Response: corrected
(7) Line 143-4. based on the figure, the region targeted by gRNA was not "close to the 5' end" as stated, it is closer to the middle of the gene sequence as shown in Figure 1C. A more accurate description would be a region in between the PAS domains.
Response: Indeed we modified the figure and the text.
(8) Line 150. The mutant allele is described as Clock1 initially, then for the rest of the paper as Clock-. SInce it is not clear that the allele is a null (see major comment #8), Clock1 should be used throughout the manuscript.
Response: the allele is named NvClk1 in the revised manuscript
(9) Figure 2A, the second CT/ZT0 is misplaced.
Response: Fig. 2 modified in the revised manuscript
(10) Figure legend for 2E and 3B. "The 1000bp upstream ATG" is unclear. I guess it means that 1000bp upstream of the putative initiation codon was used.
Response: Right, and in the revised version we analyzed 5kb upstream the putative ATG.
(11) Line 164. The authors write "We discovered..." , but wasn't it already known that these animals are behaviorally rhythmic?
Response: Fixed
(12) It would be worth mentioning in the results section the reduced amplitude of rhythms in LL compared to DD (in WT and seemingly also in Clock mutants).
Response: Indeed, we observed a significant reduction in the mean amplitude in the NvClk1-/- in DD and LL compared WT and NvClk1-/- in LD, DD and LL. However, as rhythmicity is lost by virtually all mutants in LL and DD we do not think these results add to the current interpretation of the gene function.
(13) Please correct the figure numbers in the main text, there are several mistakes.
Response: Done
(14) Line 196, most genes in the quoted study did not cycle on day 2, so whether they are truly clock controlled is questionable.
Response: We agree, identifying free-running cycling genes in cnidarian remains a challenge to overcome. One of the limitations of this study was to detect rhythmic genes in LD which conserved rhythmicity in DD. However, considering different transcriptomic studies (cited in the discussion) it seems that in the cnidaria phyla rhythmic genes in LD are not necessarily the one we identified rhythmic in DD.
(15) Line 204-206 needs to be rephrased. It is confusing.
Response: rephrased
(16) Line 216. Rephrase to something like: "A similar finding was made for."
Response: rephrased
(17) "Clock regulates genetic pathways" sounds quite odd. Do you mean it regulates preferentially specific genetic (or maybe better, molecular) pathways?
Response: rephrased
(18) Figure 4 and legend: Dashed lines indicating threshold are missing. Do the black and red dots represent WT and Clock-/-, as indicated in the legend, or up/down, as indicated in the figures?
Response: Fig.5 modified accordingly. Colors in the Volcano plot indicate Up- (black) versus Down- (red) regulated. It is now coherent within the figure.
(19) Legend for Extended figure 1. "Immature peptide sequence" is incorrect.
Response: rephrased
(20) Extended data Figure 4. What the asterisks labels is unclear.
Response: EDF4 was modified and become EDF2 with different content. The * indicates NvClk mRNA
(21) Line 228. Gene "isoforms". I guess the authors mean "paralogs".
Response: corrected.
(22) Line 232-3/Figure 3e. Please include a comparable image of the Clk ISH to facilitate the comparison of the spatial expression pattern. In addition, where and what is the "analysis" referred to - "the spatial expression pattern of Myh7 closely resembled that of Clock, as evidenced by our analysis"?
Response: the analysis has been removed from the revised manuscript because we currently cannot perform the double ish.
(23) Line 282-3. As mentioned above, it is difficult to be sure that circadian behavior is lost, if only looking at a population of animals.
Response: Fig.1 corrected
(24) Line 301-5. Rephrase.
Response: Rephrased
(25) Line 325. I am not convinced that the author can say that their mutant is amorphic. See Major comment 8.
Response: corrected.
(26) Line 351 "simplifying interactions with the environment". Please explain what is meant here.
Response: this confusing sentence has been removed from the revised manuscript