Author Response
The following is the authors’ response to the original reviews.
Response to review.
We thank the editors and reviewers for their time in assessing our manuscript. We changed the title to remove the word “all” because we realized that was hyperbolic. Corrections in response to review are in blue text throughout the manuscript document (other minor corrections are not highlighted).
eLife assessment
This study presents valuable insights into the evolution of the gasdermin family, making a strong case that a GSDMA-like gasdermin was already present in early land vertebrates and was activated by caspase-1 cleavage. Convincing biochemical evidence is provided that extant avian, reptile, and amphibian GSDMA proteins can still be activated by caspase-1 and upon cleavage induce pyroptosis-like cell death - at least in human cell lines. The caspase-1 cleavage site is only lost in mammals, which use the more recently evolved GSDMD as a caspase-1 cleavable pyroptosis inducer. The presented work will be of considerable interest to scientists working on the evolution of cell death pathways, or on cell death regulation in non-mammalian vertebrates.
We thank the editor for their time in evaluating our manuscript. We agree with the eLife assessment and with the comments of the reviewers.
Public Reviews:
Reviewer #1 (Public Review):
Summary:
The authors start out by doing a time-calibrated gene/species tree analysis of the animal gasdermin family, resulting in a dendrogram showing the relationship of the individual gasdermin subfamilies and suggesting a series of gene duplication events (and gene losses) that lead to the gasdermin distribution in extant species. They observe that the GSDMA proteins from birds, reptiles, and amphibians do not form a clade with the mammalian GSDMAs and notice that the non-mammalian GSDMA proteins share a conserved caspase-1 cleavage motif at the predicted activation site. The authors provide several series of experiments showing that the non-mammalian GSDMA proteins can indeed be activated by caspase-1 and that this activation leads to cell death (in human cells). They also investigate the role of the caspase-1 recognition tetrapeptide for cleavage by caspase-1 and for the pathogen-derived protease SpeB.
We thank the reviewer for their time in evaluating our manuscript.
Strengths:
The evolutionary analysis performed in this manuscript appears to use a broader data basis than what has been used in other published work. An interesting result of this analysis is the suggestion that GSDMA is evolutionarily older than the main mammalian pyroptotic GSDMD, and that birds, reptiles, and amphibians lack GSDMD but use GSDMA for the same purpose. The consequence that bird GSDMA should be activated by an inflammatory caspase (=caspase1) is convincingly supported by the experiments provided in the manuscript.
We thank the reviewer for their assessment of the manuscript.
Weaknesses:
- As a non-expert in phylogenetic tree reconstruction, I find the tree resulting from the authors' analysis surprising (in particular the polyphyly of GSDMA) and at odds with several other published trees of this family. The differences might be due to differences in the data being used or due to the tree construction method, but no explanation for this discrepancy is provided.
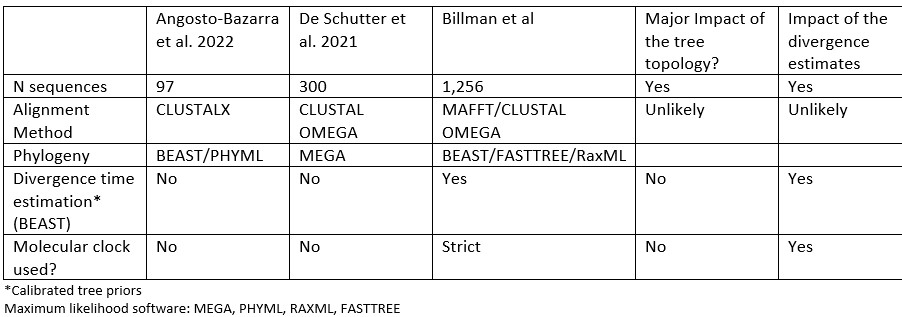
We agree, and we have modified the text to add more context to explain why our analysis generated a different topology: “In comparison to previously published studies, we used different methods to construct our gasdermin phylogenetic tree, with the result that our tree has a different topology. The topology of our tree is likely to be affected by our increased sampling of gasdermin sequences; we included 1,256 gasdermin sequences in comparison to 300 or 97 sequences used in prior studies. Prior studies used maximum likelihood tree building techniques, whereas we used a more computationally intensive Bayesian method using BEAST with strict molecular clocks that allows us to provide divergence time estimates, which we calibrated using mammal fossil estimated ages. We think that this substantially increased sampling paired with time calibration allow us to produce a more accurate phylogeny of the gasdermin protein family.”
To explain and further support our method in a more technical manner, in our phylogenetic tree, non-mammal GSDMAs are paralogous to mammals GSDMAs whereas others have found that non-mammal GSDMAs are orthologous to mammal GSDMAs. We obtained moderate support for the non-mammal GSDMA placement with Bayesian posterior 0.42 and with maximum likelihood bootstrap support of 0.96. Angosto-Bazarra et al. has for their placement a Bayesian posterior of 0.66 and maximum likelihood bootstrap support of 0.98. These are good results, but they arise from significantly fewer sequences than are included in our tree. However, in Fig S2 of Angosto-Bazarra et al. the support drops to 0.08. That the posteriors in both are not 1 indicate the presence of phylogenetic conflicts (i.e., a significant fraction of alternative trees), which means that the tree of our study or Angosto-Bazarra could be incorrect. That said, our tree is supported by biological support, and our dataset is substantially larger. To better characterize this node, further sampling with even more species would be required. We exhausted the current available sequences at the time our tree was generated.
Differences between our study and previous studies:
Author response table 1.
- While the cleavability of bird/reptile GSDMA by caspase-1 is well-supported by several experiments, the role of this cleavage for pyroptotic cell killing is addressed more superficially. One cell viability assay upon overexpression of GSDMA-NTD in human HEK293 cells is shown and one micrograph shows pyroptotic morphology upon expression in HeLa cells. It is not clear why these experiments were limited to human cells…
We did include one more experiment in human cells which is Figure 4B, in which we express full length chicken GSDMA with dimerizable caspase-1, and show that LDH release requires the cleavage site aspartate, D244. That said, we agree that our use of only human cell lines is a weakness of the paper. We thought that the best way to definitively show the interaction of caspase-1 and GSDMA was to perform experiments in chicken macrophages. Therefore, we generated a custom-raised anti-chicken-GSDMA antibody. Unfortunately, the quality of the antibody was insufficient to detect endogenous GSDMA in chicken bone marrow-derived macrophages. Off target binding prevented the observation of chicken GSDMA bands. We added a section to the discussion acknowledge the need for further studies: “In future studies, the association of bird/amphibian/reptile GSDMA and caspase-1 should be confirmed in native cells from each of these animals.”
…and why two different cell types were used for the two complementary results.
In the paper we used 293T cells and HeLa cells as generic cell types that have distinct benefits. In general, we used 293T/17 cells for experiments where high transfection efficiency was most critical, as it is simple to achieve 90% or higher transfection efficiency in this line. However, 293T/17s have poor spreading in culture and thus are not as useful for morphologic studies. 293T/17 cells do display pyroptotic ballooning upon gasdermin activation, however, the images are less pronounced in comparison to other cell types that have more distinct morphology. Therefore, we used HeLa cells for the microscopy experiments because they are more adherent and larger than 293T/17s which make for easier visualization of pyroptotic ballooning. We have added the following statement to the text to make our rationale for the use of different cell line more apparent: “In these experiments, 293T/17s were used for their high transfection efficiency, and HeLas were used for microscopy studies for their larger size and improved adherence.”
- The introduction mentions as a motivation for this work our lack of knowledge of how human GSDMA is activated. This is indeed an interesting and pressing question, but it is not really addressed in the manuscript. This is particularly true when believing the authors' dendrogram results that the bird and mammalian GSDMA families do not form a clade.
As a consequence, the significance of this finding is mostly limited to birds and reptiles.
Our aspirations were to discover hidden facets of mammal GSDMA by using a molecular evolutionary analysis. bird/amphibian/reptile GSDMA. Although we did not learn the identity of a host protease that activates mammalian GSDMA, we serendipitously discovered the evolutionary history of the association of caspase-1 with the gasdermin family. We think this manuscript provides an important and interesting advance in the field to reveal the process of evolution at work in the gasdermin family, and that the association of caspase-1 with a gasdermin to cause pyroptosis is an unbroken pairing throughout evolution. It is surprising to us that the specific gasdermin partner has changed over time.
Reviewer #2 (Public Review):
Summary:
The authors investigated the molecular evolution of members of the gasdermin (GSDM) family. By adding the evolutionary time axis of animals, they created a new molecular phylogenetic tree different from previous ones. The analyzed result verified that non-mammalian GSDMAs and mammalian GSDMAs have diverged into completely different and separate clades. Furthermore, by biochemical analyses, the authors demonstrated non-mammalian GSDMA proteins are cleaved by the host-encoded caspase-1. They also showed mammalian GSDMAs have lost the cleavage site recognized by caspase-1. Instead, the authors proposed that the newly appeared GSDMD is now cleaved by caspase-1.
We thank the reviewer for their time in evaluating our manuscript.
Through this study, we have been able to understand the changes in the molecular evolution of GSDMs, and by presenting the cleavage of GSDMAs through biochemical experiments, we have become able to grasp the comprehensive picture of this family of molecules. However, there are some parts where explanations are insufficient, so supplementary explanations and experiments seem to be necessary.
Strengths:
It has a strong impact in advancing ideas into the study of pyroptotic cell death and even inflammatory responses involving caspase-1.
We thank the reviewer for the critical consideration of the phylogeny presented.
Weaknesses:
Based on the position of mammalian GSDMA shown in the molecular phylogenetic tree (Figure 1), it may be difficult to completely agree with the authors' explanation of the evolution of GSDMA.
- Focusing on mammalian GSDMA, this group, and mammalian GSDMD diverged into two clades, and before that, GSDMA/D groups and mammalian GSDMC separated into two, more before that, GSDMB, and further before that, non-mammalian GSDMA, when we checked Figure 1. In the molecular phylogenetic tree, it is impossible that GSDMA appears during evolution again. Mammalian GSDMAs are clearly paralogous molecules to non-mammalian GSDMAs in the figure. If they are bona fide orthologous, the mammalian GSDMA group should show a sub-clade in the non-mammalian GSDMA clade. It is better to describe the plausibility of the divergence in the molecular evolution of mammalian GSDMA in the Discussion section.
We appreciate the reviewer’s careful consideration of our phylogeny. We agree that we did not make this clear enough in the discussion. Indeed, this is a confusing point, and is a critical concept in the paper. This is among our most important findings, so we have added a line addressing this finding to the abstract. We think about these concepts starting from the oldest common ancestor of a group, and then think about how genes duplicate over time. To the discussion we now begin with the following:
We discovered that GSDMA in amphibians birds and reptiles are paralogs to mammal GSDMA. Surprisingly, the GSDMA genes in both the amphibians/reptiles/birds and mammal groups appear in the exact same locus. Therefore, this GSDMA gene was present in the common ancestor of all these animals. In mammals, this GSDMA duplicated to form GSDMB and GSDMC. Finally, a new gene duplicate, GSDMD, arose in a different chromosomal location. Then this GSDMD gene became a superior target for caspase-1 after developing the exosite. Once GSDMD had evolved, we speculate that the mammalian GSDMA became a pseudogene that was available to evolve a new function. This new function included a new promoter to express mammalian GSDMA primarily in the skin, and perhaps acquisition of a new host protease that has yet to be discovered.
In further support of the topology of our Bayesian tree in Figure 1, we also performed a maximum likelihood analysis, which also placed the GSDMA genes into similarly distinct clades (Figure 1-S3). Finally, we have biological evidence to support this reasoning, where caspase-1 cleaves non-mammal GSDMAs and also mammal GSDMD (and no longer can cleave mammal GSDMA).
- Regarding (1), it is recommended that the authors reconsider the validity of estimates of divergence dates by focusing on mammalian species divergence. Because the validity of this estimation requires a recheck of the molecular phylogenetic tree, including alignment.
Our reconstructed evolution of gasdermins is consistent with the mammal tree of life. We constrained Bayesian estimation of divergences using soft calibrations from mammal fossil estimated ages. We have included the fossil calibration of mammalian gasdermins to the results section and to our methods.
- If GSDMB and/or GSDMC between non-mammalian GSDMA and mammalian GSDMD as shown in the molecular phylogenetic tree would be cleaved by caspase-1, the story of this study becomes clearer. The authors should try that possibility.
It is known that mammal GSDMB and GSDMC cannot be activated by caspase-1. We propose that GSDMA was cleaved by caspase-1 only in extinct mammals that had not yet associated GSDMD with caspase-1. Such an extinct mammal could have encoded a GSDMA cleaved by caspase-1, a GSDMB cleaved by granzyme A, and GDSMC cleaved by caspase-8. Later, the GSDMA gene was again duplicated to form GSDMD. After GSDMD was targeted by caspase-1, then GSDMA was free to gain its current function in barrier tissues.
Reviewer #1 (Recommendations For The Authors):
As a non-expert on phylogenetic tree construction, I found the "time-calibrated maximum clade credibility coalescent tree" hard to digest. I would have liked to see an explanation of how this method is different from what has been used before and why the authors consider it to be better. This is particularly important when considering that the resulting tree shown in Figure 1 is quite different from other published trees of the same family (e.g. https://www.ncbi.nlm.nih.gov/pmc/articles/PMC8742441 where the GSDMA family appears monophyletic).
Please see response to Reviewer 1 weaknesses above. Also, we have moved the text “time-calibrated maximum clade credibility coalescent tree” to the figure legend.
In the bioinformatical analysis of the conserved caspase-1 cleavage motif in bird GSDMA sequences, I would recommend also addressing the residue behind the cleavage site Asp, as this position has an unusually high conservation (mostly Gly) in bird GSDMA.
This is a great observation. We suspect that this may reflect a need for flexibility in the secondary structure to allow the cleavage site to enter the enzymatic pocket of the caspase. This residue is also similarly enriched in mammal GSDMD, which is also cleaved by caspase-1. We also note high conservation of a P2' proline residue in birds with the FASD tetrapeptide, which could also be important for displaying the tetrapeptide to the caspase.
This comment prompted us to search the literature for evidence of these residues in caspase-1 substrate preference studies. Remarkably, a P1' glycine and P2` proline are among the most enriched residues in human caspase-1 targets. This supports our hypothesis that caspase-1 cleaves GSDMA in non-mammals. We added the following to the results section: “Additionally, the P1' residue in amphibian, bird and reptile GSDMA was often a glycine, and the P2' residue was often a proline, especially in birds with FASD/FVSD tetrapeptides (Fig. 2B). A small P1' residue is preferred by all caspases. By using a peptide library, glycine has been determined to be the optimal P1' residue for caspase-1 and caspase-4. Further, in a review of the natural substrates of caspase-1, glycine was the second most common P1' residue, and proline was the most common P2' residue. These preferences were not observed for caspase-9.”
Finally, I would like the authors to at least explain why the cell viability assays were done in 293T cells while the micrographs were done in HeLa cells. Why not show both experiments for both cell types?
In the paper we used 293T cells and HeLa cells as generic cell types that have distinct benefits. In general, we used 293T/17 cells for experiments where high transfection efficiency was most critical, as it is simple to achieve 90% or higher transfection efficiency in this line. However, 293T cells have poor spreading in culture and thus are not as useful for morphologic studies. 293T/17 cells do display pyroptotic ballooning upon gasdermin activation, however, the images are less pronounced in comparison to other cell types that have more distinct morphology. Therefore, we used HeLa cells for the microscopy experiments because they are more adherent and larger than 293T/17s which make for easier visualization of pyroptotic ballooning. We have added the following statement to the text to make our rationale for the use of different cell line more apparent: “In these experiments, 293T/17s were used for their high transfection efficiency, and HeLas were used for microscopy studies for their larger size and improved adherence.”
There are a number of minor points related to language and presentation:
- the expressions "pathogens contaminate the cytosol", "mammals can encode..", "an outsized effect" are unusual and might be rephrased.
We changed these to:
“manipulate the host cell, sometimes contaminating the cytosol with pathogen associated molecular patterns, or disrupting aspects of normal cell physiology”,
“Only mammals encode GSDMC and GSDMD alongside the other four gasdermins.”,
and
“greater effect”
- in line 87 the abbreviation "GSDMEc" is first used without explanation (of the "c").
This is an important distinction, as GSDMEc proteins were only recently uncovered. To remedy this, we have added the following text following line 87: “This gasdermin was recently identified as an ortholog of GSDMA.
It was called GSDMEc, following the nomenclature of other duplications of GSDME in bony fish that have been named GSDMEa and GSDMEb.”
Corrected
- line 186ff the sentence "We believe..." does not appear to make sense.
We revised the text to make this clear, changing the text to now read “We hypothesized that activating pyroptosis using separate gasdermins for caspase-1 and caspase-3 is a useful adaptation and allows for fine-tuning of these separate pathways. In mammals, this separation depends on the activation of GSDMD by caspase-1 and the activation of GSDME by caspase-3.”
- many figures use pictures rather than text to represent species groups. These pictures are not always intuitive. As an example, in Figure 6 the 'snake' represents amphibians. After reading the text, I understand that these should probably be the caecilian amphibians, but not every reader might know what these critters look like. In Figure 7, I have no idea what the black blob (2nd image from top) is supposed to be.
In crafting the manuscript, we found the use of text to denote the various species to be cumbersome. The species silhouettes are a standard graphical depiction used in evolutionary biology, which we think aids readability to the figures. For example, in a paper cited in our manuscript, these same silhouettes were used to depict the evolution of GSDMs (https://doi.org/10.3389/fcell.2022.952015 Figure 1A, Figure 3D, Figure 4G). However, we agree that many readers will not know that caecilians are legless amphibians that resemble snakes in their body morphology, but are not close to snakes by phylogeny. We think it is important to use an image of a caecilian amphibian because the more iconic amphibians (frogs, salamanders) do not encode GSDMA. To increase clarity, we have mentioned the morphology of caecilians in the legend of Figure 2, Figure 6, and Figure 7 when caecilican amphibians are first introduced.
In Figure 2: “Note, that caecilians morphologically are similar to snakes in their lack of legs and elongated body, however, this is an example of convergent evolution as caecilians are amphibians and are thus more closely related to frogs and salamanders than snakes.”
In Figure 6: “M. unicolor is an amphibian despite sharing morphological similarity to a snake.”
In Figure 7: “In caecilian amphibians, which are morphologically similar to snakes, birds, and reptiles, GSDMA is cleaved by caspase-1.”
The black blob is the mollusk Lingula anatina, which unfortunately has an indistinct silhouette. To clarify this, we have added text to label the images in Figure 7.
Reviewer #2 (Recommendations For The Authors):
- Line 214, in "(Fig. 3-S2) Human and mouse ..", it is necessary to type a period.
- Line 238, in the subtitle, GSMA should be amended to GSDMA.
These have both been corrected.




