Peer review process
Not revised: This Reviewed Preprint includes the authors’ original preprint (without revision), an eLife assessment, public reviews, and a provisional response from the authors.
Read more about eLife’s peer review process.Editors
- Reviewing EditorHaifan LinYale University, New Haven, United States of America
- Senior EditorUtpal BanerjeeUniversity of California, Los Angeles, Los Angeles, United States of America
Reviewer #1 (Public Review):
Summary:
Here, the authors, Barber AG et al, developed a new mouse model and investigated an importance of Musashi-2 in lung cancer. Specifically, they found that Musashi-2 is important for lung cancer cells as it controls cancer cell growth, and also regulates several genes that also control cancer cell growth. Development of a new Musashi-2 mouse model is a plus, which confirmed Musashi-2 importance for lung cancer survival, and finding several genes that Musashi controls that are important for lung cancer growth. Additionally, they demonstrated that Musashi-2 overexpression which is tracked by GFP is preferred in lung adenocarcinoma cells. The data is rigorous and only minor revisions are requested.
Strengths:
Authors achieved their goals, by developing new Musashi-2 mouse model, confirming Musashi-2 importance for lung cancer survival, and finding several genes that Musashi controls that are important for lung cancer growth.
Weaknesses:
The findings of Musashi-2 mouse and human lung cancer growth control are not that novel as prior publication in 2016 showed that already, again, in both human and mouse models (Kudinov et al PNAS, PMID: 27274057), and also the authors missed the point of that paper which did use both miuse and human models to show impact on inbvasion and metastasis- both in vitro and in vivo. Additionally, another publication is currently under revisions recently also generated new Musashi-2 transgenic mouse model which confirmed Musashi-2 support of lung cancer growth (Bychkov I et al, PMID: 37398283; https://www.biorxiv.org/content/10.1101/2023.06.13.544756v1). Another weakness is that Musashi-2 cannot be effectively targeted and the new genes the authors found that Musashi-2 regulates are likely to be also difficult therapeutic targets. Therefore, impact of this new investigation is relatively modest in the field.
Major suggestions:
(1) Figure 3: it is unclear what is the efficiency of Msi2 deletion shRNA - could you demonstrate it by at least two independent methods? (QPCR, Western, or IHC?) please quantitate the data.
(2) In Figure 4, similarly, it is unclear if Msi2 depletion was effective- and what is shRNA efficiency. Please test this by at least two independent methods (QPCR, Western, or IHC) and also please quantitate the data
(3) the reason for impairment of cell growth demonstrated in Figs 3 and 4 is not clear: is it apoptosis? Necrosis? Cell cycle defects? Autophagy? Senescence? Please probe 2-3 possibilities and provide the data.
(4) Since Musashi-1 is a Musashi-2 paralogue that could compensate for Musashi-2 loss, please test Msi1 expression levels in matching Fig 3 and Fig 4 sections (in cells/ tumors with Msi2 deletion and in KP cells with Msi2 shRNA). One method could suffice here.
(5) It is not exactly clear why RNA-seq (as opposed to proteomics) was done to investigate downstream Msi2 targets (since Msi2 is in first place, translational and not transcriptional regulator)- . RNA effects in Fig 5J are quite modest, 2-fold or so. It would be useful (if antibodies available) to test four targets in Fig 5J by Western blot, to see any impact of musashi-2 depletion on those target protein levels. Indeed, several papers - including Kudinov et al PNAS, PMID: 27274057, Makhov P et al PMID: 33723247 and PMID: 37173995 - used proteomics/ RIP approaches and found direct Musashi-2 targets in lung cancer, including EGFR, and others.
Reviewer #2 (Public Review):
Summary:
Alison G. Barber et al. reports the function of Msi2 in mouse models of non-small cell lung cancer. The expression of Msi2 in normal lung was evaluated using a knockin reporter allele. Msi2 expressing cells were found to be around 30-40% in normal lung epithelium without a strong bias in subsets of lung cells. Knocking out Msi2 in a KrasG12D and P53 KO model reduced lung cancer initiation. Knocking down Msi2 in established lung cancer cells reduced in vitro sphere formation and in vivo xenograft. Finally, the authors identified several genes whose expression was downregulated by Msi2 knockdown. Knocking down four of these genes, including Ptgds, Arl2bp, hRnf157, and Syt11, each with a single shRNA, reduced lung sphere formation in vitro, suggesting their involvement in lung cancer.
Strengths:
This manuscript represents an interesting advance on the role of Msi2 in lung cancer. While some of the data (for example the knockdown effect of Msi2 in established lung cancer cells) corroborated previous findings, the study of Msi2 expression in normal lung and the characterization of the KO phenotype in lung cancer initiation are new and interesting.
Weaknesses:
Two areas can be further strengthened. Several conclusions are not fully supported by the existing data. The stable/dynamic nature of Msi2 expressing cells in lung would benefit from more detailed investigations for proper data interpretation.
(1) It will be interesting to determine whether Msi2+ cells are a relatively stable subset or rather the Msi2+ cells in lung is a dynamic concept that is transient or interconvertible. This is relevant to the interpretation of what Msi2 positivity really means.
(2) Does Kras mutation and/or p53 loss upregulate Msi2? This point and the point above are related to whether Msi2+ cells are truly more susceptible to tumorigenesis, as the authors suggested.
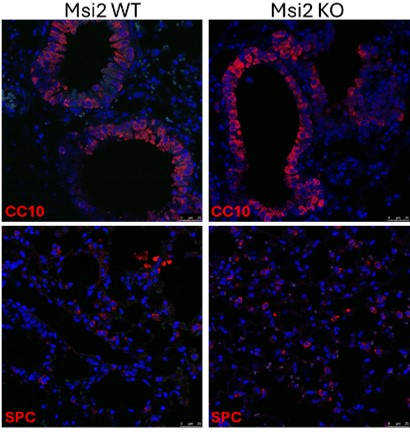
(3) The KO of Msi2 reducing tumor number and burden in the lung cancer initiation model is interesting. However, there are two alternative interpretations. First, it is possible that the Msi2 KO mice (without Kras activation and p53 loss) has reduced total lung cell numbers or altered percentage of stem cells. There is currently only one sentence citing data not shown on line 125, commenting that there is no difference in BASC and AT2 cell populations. It will be helpful that such data are shown and the effect of KO on overall lung mass or cellularity is clarified. Second, the phenotype may also be due to a difference in the efficiencies of cre on Kras and p53 in the Msi2 WT and KO mice.
(4) All shRNA experiments (for both Msi2 KD and the KD of candidate genes) utilized a single shRNA. This approach cannot exclude off-target effects of the shRNA.
(5) The technical details of the PDX experiment (Figure 4F) are not fully explained.
Reviewer #3 (Public Review):
Summary:
In this manuscript, Barber and colleagues propose a dual role for the RNA-binding protein Mushashi-2 (Msi2) in lung adenocarcinoma initial transformation and subsequent tumor propagation. First, authors show that Msi2 is expressed in a subset of Club/BASC (37%) and AT2 (26%) cells in the normal lung and displayed a distinct transcriptional profile than non-expressing Msi2 cells. Furthermore, Msi2 is broadly expressed/activated in vivo in genetically induced lung adenocarcinoma tumors (Kras/p53 mouse model) and Msi2+ cells displayed a significantly higher ability to form tumor spheres in vitro. Authors demonstrated by in vivo and in vitro assays that Msi2 loss of function significantly impair tumor growth and progression in lung adenocarcinoma. Data showed that Msi2 function is conserved in human adenocarcinoma tumor growth in patient-derived xenograft. Lastly, novel genes regulated by Msi2 and involved in lung adenocarcinoma tumor growth were identified.
Strengths:
The authors provided convincing data for a key role of Msi2 in lung adenocarcinoma tumor progression and growth. Multiple evidences using Msi2 knock-out genetic mouse model and shRNA knock-down in tumor sphere formation assay are clearly demonstrated. The conservation and importance of Msi2 was further shown in human patient-derived xenograft. Although specific cell types (Club/BASC, AT2) were not isolated, authors further delved in the transcriptional difference between Msi2+ and Msi2- cells in the normal lung. Furthermore, novel genes and pathways regulated by Msi2 in lung adenocarcinoma were identified and tested for their ability to inhibit tumor growth in vitro. These 2 RNA-Seq datasets will be useful in the future and provide a basis to explore 1) potential propensity of a given cell to initiate oncogenic transformation, and 2) potential novel regulators of lung adenocarcinoma.
Weaknesses:
Although this work strongly demonstrated the importance of Msi2 in lung adenocarcinoma tumor progression and growth, the following points remain to be clarified or addressed.
- In Figure 1, characterization of Msi2 expression in the normal mouse lung was carried out by using a Msi2-GFP Knock-in reporter and analyzed by flow cytometry followed by cytospins and immunostaining. Additional characterization of Msi2 expression by co-immunostaining with well-known markers of airway and alveolar cell types in intact lung tissue will strengthen the existing data and provide more specific information about Msi2 expression and abundancy in relevant cell types. It will be also interesting to know whether Msi2 is expressed or not in other abundant lung cell types such as ciliated and AT1 cells.
- While this set of experiments provide strong evidence that Msi2 is required for tumor progression and growth in lung adenocarcinoma, it is unclear whether normal Msi2+ lung cells are more responsive to transformation or whether Msi2 is upregulated early during the process of tumorigenesis. Future lineage tracing experiments using Msi2-CreER and mouse models of chemically-induced lung carcinogenesis will provide additional data that will fully support this claim.
- In Figure 4F, Patient-derived xenograft (PDX) assays were conducted in 2 patients only and the percentage of cells infected by shRNA-Msi2 is low in both PDX (30% and 10% for patient 1 and 2 respectively). It is surprising that Msi2 downregulation in a small percentage of tumor cells has such a dramatic effect on tumor growth and expansion. Confirmation of this finding with additional patient samples would suggest an important non-cell autonomous role for Msi2 in lung adenocarcinoma.