Author response:
The following is the authors’ response to the original reviews.
Public Reviews:
Reviewer #1 (Public review):
Summary:
The manuscript entitled "Phosphodiesterase 1A Physically Interacts with YTHDF2 and Reinforces the Progression of Non-Small Cell Lung Cancer" explores the role of PDE1A in promoting NSCLC progression by binding to the m6A reader YTHDF2 and regulating the mRNA stability of several novel target genes, consequently activating the STAT3 pathway and leading to metastasis and drug resistance.
Strengths:
The study addresses a novel mechanism involving PDE1A and YTHDF2 interaction in NSCLC, contributing to our understanding of cancer progression.
Weaknesses:
The following issues should be addressed:
(1) The body weight changes and/or survival times of each group in the in vivo metastasis studies should be provided.
Thank you for your suggestion! We have already provided the body weight of each group in the in vivo metastasis studies in FigureS4D and FigureS5D (see below).
(2) In Figure 7, the direct binding between YTHDF2 and the potential target genes should be further validated by silencing YTHDF2 to observe the half-life of the mRNA levels of target genes, in addition to silencing PDE1A.
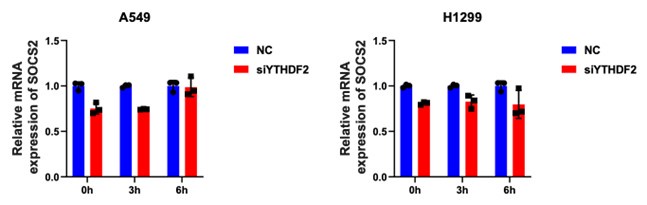
Thank you for your suggestion! We have found that siYTHDF2 does not significantly affect expression of SOCS2 in NSCLC cells (see author response image 1 below). We hypothesize that YTHDF2 functions as a m6A reader to recognize the target mRNA, thus if YTHDF2 is silence by siRNA, there is still some expression in the cells, allowing it to continue recognizing and exerting its function. Therefore, the mRNA of SOCS2 could not significantly affect expressed. However, PDE1A functions as a degrader of mRNA, thus when it is disrupted, the mRNA degradation effect could be strong.
Author response image 1.
SOCS2 mRNA expression after siYTHDF2 in NSCLC cells
(3) In Figure 7, the potential methylation sites of "A" on the target genes such as SOCS2 should be verified by mutation analysis, followed by m6A IP or reporter assays.
Thank you for your suggestion! The m6A IP or reporter assays may be carried out to detect the potential methylation sites in future. We have added the suggestion in manuscript “Meanwhile, YTHDF2 might act as an m6A RNA “reader” by interacting with PDE1A, but the mechanism might need further investigation”.
(4) In Figure 6G, the correlation between the mRNA levels of STAT3 and YTHDF2 needs clarification. According to the authors' mechanism, the STAT3 pathway is activated, rather than upregulation of mRNA levels (or protein levels, as shown in Figure 6F). Figure 7 does not provide evidence that STAT3 is a bona fide target gene regulated by YTHDF2.
Thank you for your suggestion! The reviewer is right, STAT3 pathway is activated, rather than upregulation of mRNA levels by YTHDF2, so the relationship between YTHDF2 mRNA and STAT3 mRNA is not suitable for this study. Meanwhile, the relationship between YTHDF2 mRNA and STAT3 mRNA is not as strong as we expected with Pearson value 0.37. Thus, we have already deleted Figure 6G in the revised version.
(5) The final figure, which discusses sensitization to cisplatin by PDE1A suppression, does not appear to be closely related to the interaction or regulation of PDE1A/YTHDF2. If the authors claim this is an m6A-associated event, additional evidence is needed. Otherwise, this part could be removed from the manuscript.
Thank you for your suggestion! We have already deleted Figure 8 just as the reviewer suggested.
Reviewer #2 (Public review):
This manuscript aims to investigate the biological impact and mechanisms of phosphodiesterase 1A (PDE1A) in promoting non-small cell lung cancer (NSCLC) progression. They first analyzed several databases and used three established NSCLC cell lines and a normal cell line to demonstrate that PDE1A is overexpressed in lung cancer and its expression negatively correlated with the outcomes of patients. Based on this data, they suggested PDE1A could be considered as a novel prognostic predictor in lung cancer treatment and progression. To study the biological function of PDE1A in NSCLC, they focused on testing the effect of inhibition of PDE1A genetically and pharmacologically on cell proliferation, migration, and invasion in vitro. They also used an experimental metastasis model via tail vein injection of H1299 cells to test if PDE1A promoted metastasis. By database analysis, they also decided to investigate if PDE1A promoted angiogenesis by co-culturing NSCLC cells with HUVECs as well as assessing the tumors from the subcutaneous xenograft model. However, in this model, whether PDE1A modulation impacted tumor metastasis was not examined. To address the mechanism of how PDE1A promotes metastasis, the authors again performed a bioinformatic and GSEA enrichment analysis and confirmed PDE1A indeed activated STAT3 signaling to promote migration. In combination with IP followed by Mass spectrometry, they found PDE1A is a partner of YTHDF2, the cooperation of PDE1A and YTHDF2 negatively regulated SOCS2 mRNA as demonstrated by RIP assay, and ultimately activated STAT3 signaling. Finally, the authors shifted the direction from metastasis to chemoresistance, specifically, they found that PDEA1 inhibitions sensitized NSCLC cells to cisplatin through MET and NRF2 signaling.
Strength:
Overall, the manuscript was well-written and the majority of the data supported the conclusions. The authors used a series of methods including cell lines, animal models, and database analysis to demonstrate the novel roles and mechanism of how PDE1 promotes NSCLC invasion and metastasis as well as cisplatin sensitivity. Given that PDE1A inhibitors have been perused to use in clinic, this study provided valuable findings that have the translational potential for NSCLC treatment.
Weaknesses:
The role of YTHDF2 in PDE1A-promoted tumor metastasis was not investigated. To make the findings more clinical and physiologically relevant, it would be interesting to test if inhibition of PDE1A impacts metastasis using lung cancer orthotopic and patient-derived xenograft models. It is also important to use a cisplatin-resistant NSCLC cell line to test if a PDE1A inhibitor has the potential to sensitize cisplatin in vitro and in vivo.
Thank you for your suggestion! The role of YTHDF2 in PDE1A-promoted tumor metastasis may need in vivo analysis. Therefore, we discussed the point in the discussion section “In addition, it is worth testing if PDE1A inhibition affects metastasis in lung cancer orthotopic and patient-derived xenograft models. The role of YTHDF2 in PDE1A-driven tumor metastasis should be elucidated in future studies”.
The reviewer is absolutely right, it is very important to use a cisplatin-resistant NSCLC cell line to test the potential effect of PDE1A in sensitization to cisplatin. The current data could not support the conclusion, more data is needed to make the final conclusion. As suggested by reviewer 1, we have deleted these data in this version.
Furthermore, this study relied heavily on different database analyses, although providing novel and compelling data that was followed up and confirmed in the paper, it is critical to have detailed statistical description section on data acquisition throughout the manuscript.
Thank you for your suggestion! We have already added the detailed statistical description section in Figure legends.
Recommendations for the authors:
Reviewer #1 (Recommendations for the authors):
(1) Scale Bar Display: Scale bars should be included in Figures 4F, 5F, and 6E to ensure clarity and accuracy in the presented microscopic images.
Thank you for your suggestion! We have already added the scale bars on Figures 4F, 5F, and 6E.
(2) HE Staining Images: The authors are suggested to provide more images for HE staining of lungs to offer a comprehensive visual representation and to substantiate the findings.
Thank you for your suggestion! We have already provided more images for HE staining of lungs in Figure S4E and Figure S5E.
Reviewer #2 (Recommendations for the authors):
It would be helpful to clarify several points in the manuscript for better understanding.
(1)The HELF cells were stated between the epithelial cell line (page 7, line 118) and fibroblast (page 12, line 288) which needs to be clarified. It is not clear if the cells used in this study were periodically authenticated.
Thank you for your suggestion! We have already revised the expression of HELF cells, and it is actually the human lung fibroblasts.
(2) More details could be added to the methods such as the amount of Matrigel coated for invasion assay and the components for the lysis buffer and IP buffer.
Thank you for your suggestion! We have already added more details in the Methods section.
(3) Providing the rationale for using 20% FBS instead of using some chemoattracts such as EGF, LPA, or HGF or a low level of FBS for migration will be helpful.
Thank you for your suggestion! Although chemoattracts are suitable for cell migration experiment, and 20% FBS is also suitable for cell migration experiment. We listed the literatures using this system below for example.
(1) Xiaolin Peng, Zhengming Wang, Yang Liu. et al. Oxyfadichalcone C inhibits melanoma A375 cell proliferation and metastasis via suppressing PI3K/Akt and MAPK/ERK pathways, Life Sciences, 2018, 206, 35-44. https://doi.org/10.1016/j.lfs.2018.05.032
(2) Rong, S., Dai, B., Yang, C. et al. HNRNPC modulates PKM alternative splicing via m6A methylation, upregulating PKM2 expression to promote aerobic glycolysis in papillary thyroid carcinoma and drive malignant progression. J Transl Med, 2024, 22, 914 (2024). https://doi.org/10.1186/s12967-024-05668-9
(4) For HPA analysis In Figure 1, it would be great to assess how many lung cancer cases are NSCLC and define IDO/area for the y-axis.
Thank you for your suggestion! There are 19 samples were analyzed, they are all NSCLC sample, and we have already revised our manuscript accordingly. Meanwhile, we also made a mistake, it should be IOD/area which means Integral optical density/area. We have revised the Figures and Figure legends.
(5) On page 23, line 480, "Therefore, this study reveals the effect and mechanism of PDEA1 in promoting HCC metastasis...", should HCC be NSCLC?
Thank you for your suggestion! We have already revised the manuscript accordingly.
(6) Specific scramble siRNAs should be clearly shown in their respective figures. In Figure 7F, it is not clear why DMSO did not scramble siRNA was used as the control.
Thank you for your suggestion! It is our fault to show the DMSO in Figure 5F, DMSO is the negative control of Figure 5G, and we have revised the Figure 5F and 5G accordingly.




