The AFF4 scaffold binds human P-TEFb adjacent to HIV Tat
Figures
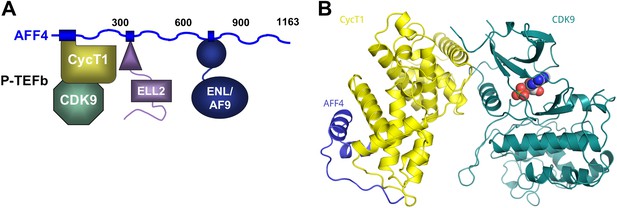
AFF4 binds CycT1 distal to CDK9.
(A) Schematic model of the SEC. AFF4 is an intrinsically disordered scaffold that binds partners via 20–50 residue segments. (B) Ribbon diagram showing the strand–helix–helix arrangement of AFF4 (blue) bound to CycT1 (yellow) remote from CDK9 (teal). AFF4 adopts an extended conformation with no intramolecular tertiary contacts. AMPPNP (spheres) is bound to CDK9.
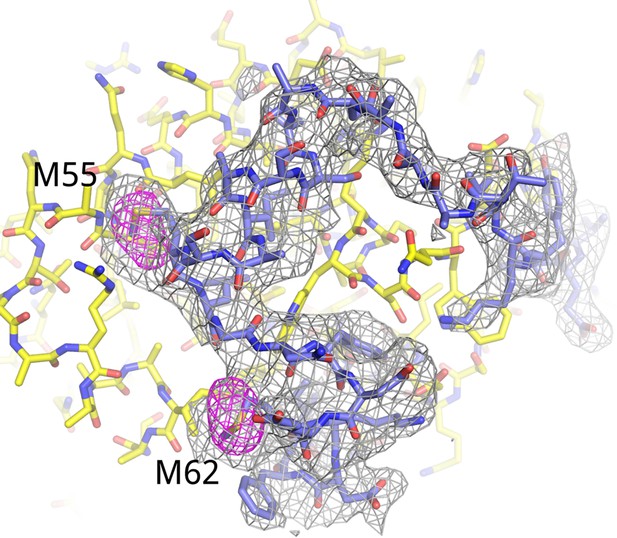
Electron density for AFF42–73.
Fo-Fc omit map (3.5 σ, gray) for residues 34–66 of AFF4. Anomalous difference map (5 σ, red) shows the positions of the methionine residues in AFF4. The anomalous difference Fourier was calculated using X-ray data recorded from a crystal grown with Seleno-methionine labeled AFF4.
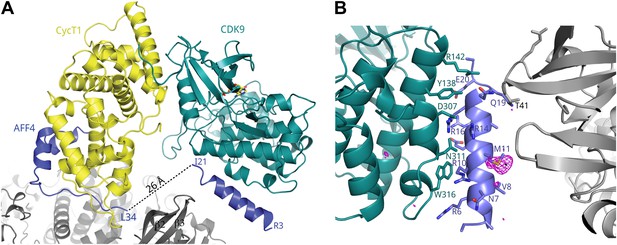
A crystal contact formed by AFF42–21.
(A) An isolated helix from the aff4 N-terminus, packs loosely against αE and αI of one CDK9 subunit (chain C) and makes contacts with the β2-β3 loop of a symmetry-related CDK9 molecule. (B) Interactions between CDK9 residues (teal sticks) and the isolated aff4 helix H0 (blue). Anomalous difference map (3.5 σ, red) shows the position of SeMet11. The occurrence of this helix in only one of three independent complexes in the crystals and the lack of electrostatic complementarity with CDK9 suggest that the helix may depend on the crystal environment and the kinase subunit is not the functional partner of this sequence.
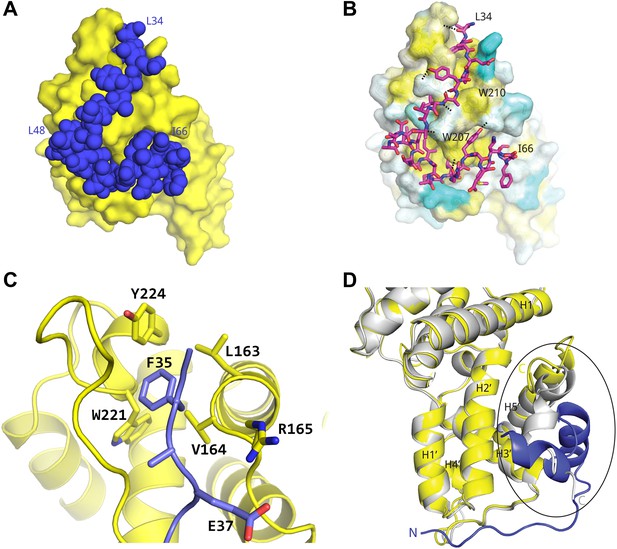
Basis for AFF4 scaffold recognition by P-TEFb.
(A) AFF4 residues 34–66 (blue spheres) fill grooves on CycT1 (yellow surface). (B) Chemical complementarity mediates AFF4 binding. Exposed hydrophobic residues of CycT1 (yellow surface) are buried in the AFF4 complex. Hydrogen bonds (black dotted lines) also mediate binding. (C) AFF4 Phe35 is buried in a hydrophobic pocket formed by aromatic and nonpolar residues on the surface of CycT1. (D) The C-terminus of the CycT1 cyclin domain (gray in the ellipse) adjusts to make contacts with AFF4 (blue).
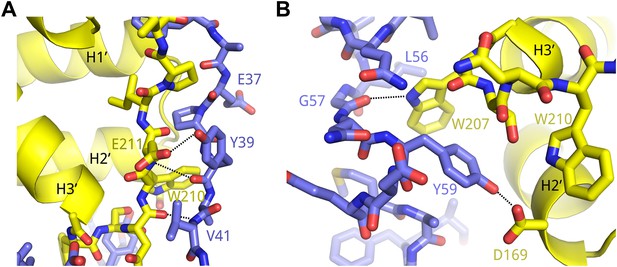
Example interactions between AFF4 and P-TEFb.
(A) Main-chain hydrogen bonds between the extended aff4 peptide 34–41 and CycT1. (B) Hydrophobic interactions with CycT1 Trp207 and hydrogen bonds of CycT1 Trp207 and Asp169 anchor the aff4 α1–α2 loop.
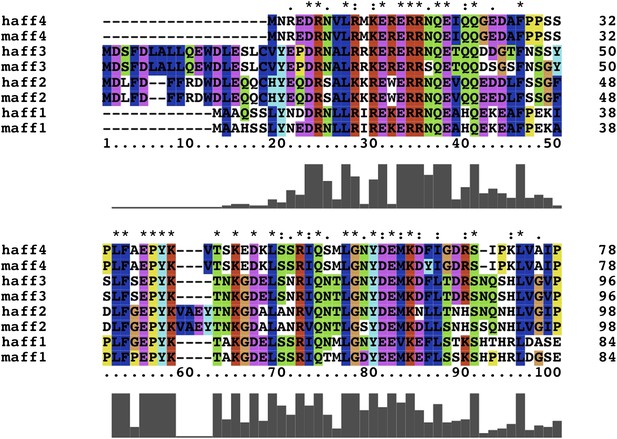
Conserved AFF4 sequences mediate P-TEFb recognition.
Multiple sequence alignment of amino acids 1–78 (AFF4 numbering) for human and mouse AF4 family members.
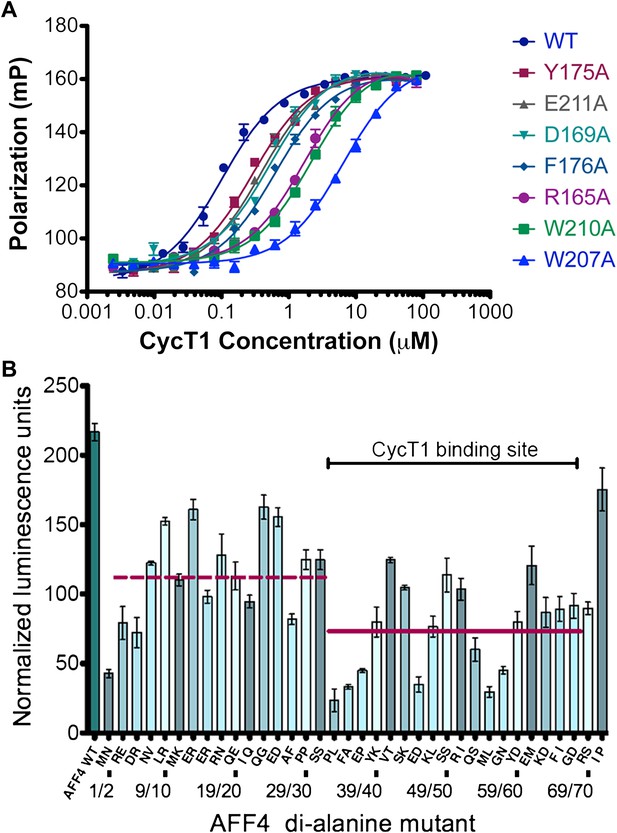
AFF4 interface mediates P-TEFb recognition.
(A) Mutations of CycT1 contact residues reduce AFF4 affinity. Fluorescence polarization of fluorescein-labeled AFF432–67 (5 nM) is plotted as a function of the concentration of the indicated CycT1 variant. (B) Transcriptional effects of AFF4 tandem Ala mutants. Stimulation of Tat-independent transcription from the HIV LTR was measured in extracts of cells cotransfected with a luciferase reporter construct and an expression vector for the indicated di-Ala AFF4 variant. Activity was normalized to the level of AFF4 expression. Values represent the mean of three independent assays. Tandem alanine substitutions cover the first 72 residues of AFF4. Horizontal lines correspond to the mean stimulation of di-Ala substitutions in residues 3–32 (left; 113.9 ± 5.1) and 33–66 (right; 73.6 ± 4.9).
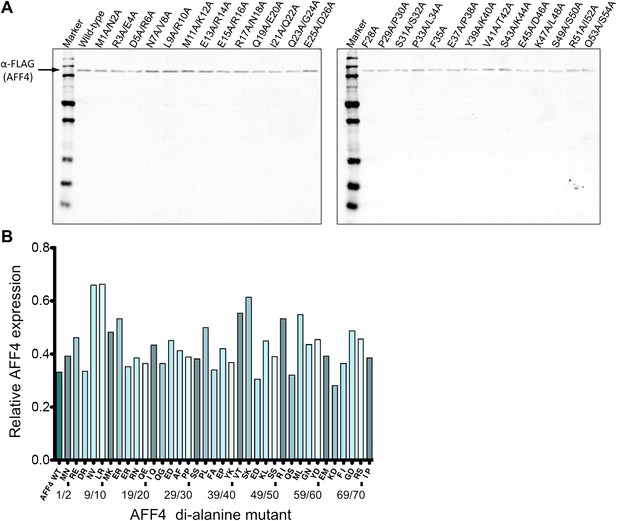
Expression levels of AFF4 variants.
(A) Representative Western blot probed with anti-FLAG antibodies to measure the level of FLAG-tagged aff4 variants in HeLa cell lysates. (B) Expression levels of aff4 variants. Values are the average of three biological replicates. The tandem alanine mutants showed similar levels of expression.
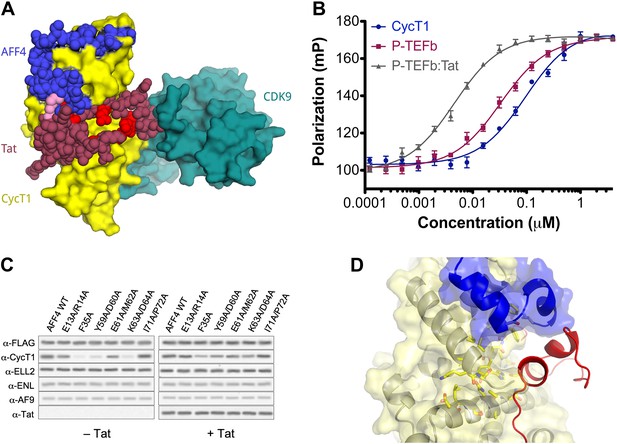
AFF4 binds in position to make direct contacts with HIV-1 Tat.
(A) Superposition of the AFF4-P-TEFb complex and the Tat-P-TEFb complex using the cyclin subunit (yellow) shows the close proximity of AFF4 (blue) and Tat (red). Tat Lys28 (pink), where acetylation stimulates function, as well as other residues essential for Tat transcriptional activation (D'Orso et al., 2012) that are exposed to solvent in the Tat-P-TEFb complex (bright red) are positioned adjacent to AFF4. (B) Tat enhances AFF4 binding in vitro. Fluorescence polarization of fluorescein-labeled AFF432–67 (5 nM) is plotted as a function of the concentration of CycT1 (blue circles), P-TEFb (red squares), and Tat-P-TEFb (green triangles). (C) Alanine substitutions in the P-TEFb binding site of AFF4 reduce CycT1 binding but not associations of other SEC subunits in HeLa cells. Western blots show associations of each indicated factor with different FLAG-tagged AFF4 variants (top). Lysates were immunoprecipitated with an anti-FLAG antibody. Expression of Tat (right) rescues defects in CycT1 binding, except for the E61/M62 double alanine mutant. This mutant in the predicted AFF4-Tat interface shows equal small defects in P-TEFb binding in the absence (left) and presence (right) of Tat. (D) AFF4 (blue) and CycT1 (yellow) create an intersubunit pocket where Tat (red) can bind with minor structural adjustments. The program DoGSiteScorer (Volkamer et al., 2012) assigns this cleft a high druggability score (0.83 out of 0–1.0) and shows that it contains the most nonpolar surface of any pocket in the AFF4-P-TEFb structure.
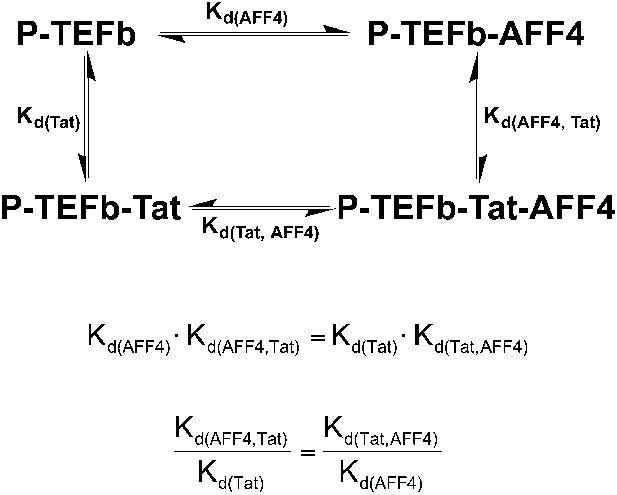
Thermodynamic cycle for AFF4 and Tat binding to P-TEFb.
The enhancement of Tat affinity for P-TEFb by aff4 is equal to the ratio of aff4 affinity for P-TEFb in the presence and absence of Tat.
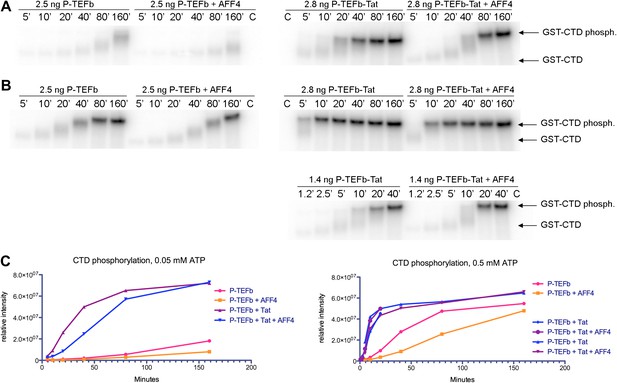
Kinase activity of P-TEFb and P-TEFb-Tat complexes with AFF4.
(A) Autoradiogram showing phosphorylation of GST-CTD (500 ng) by P-TEFb and P-TEFb-Tat with and without excess (0.28 μM) AFF42-73 in the presence of low (50 μM) ATP. (B) Phosphorylation of 500 ng GST-CTD by P-TEFb and P-TEFb-Tat with and without excess (0.28 μM) AFF42–73 in the presence of saturating (500 μM) ATP. AFF4 reduces the activity of P-TEFb twofold and has little influence on the kinase activity of Tat-P-TEFb. Tat-P-TEFb is, however, sevenfold to tenfold more active than P-TEFb. Lane 3 in panels (A and B) is a control without GST-CTD. (C) Quantitation of the radioactive GST-CTD in (A and B).
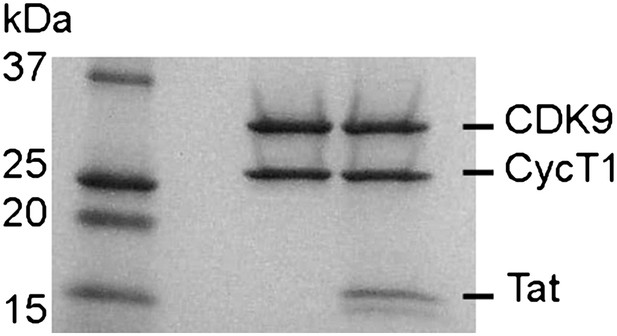
SDS polyacrylamide gel of P-TEFb and P-TEFb-Tat at the same ratio as they were used in the kinase assay.
https://doi.org/10.7554/eLife.00327.017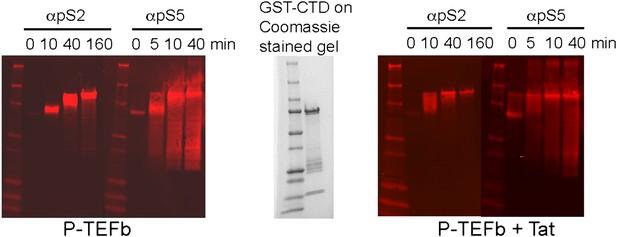
Western blots of kinase reaction products from panel B. Phosphorylated CTD was detected with anti-phoshoSer2 and anti-phoshoSer5 antibodies.
The GST-CTD was phosphorylated on Ser2 and Ser5. However, the Ser2 phosphorylation was detected only on the full-length CTD, while Ser5 phosphorylation was detected disproportionately on proteolytic fragments of the CTD compared to the full-length CTD domain.
Tables
X-ray data collection and refinement statistics for AFF4-P-TEFb-AMPPNP
| Data collection | AFF4-P-TEFb-AMPPNP |
| Space group | P212121 |
| Cell dimensions: a, b, c | 100.691, 126.298, 195.626 |
| Resolution (Å)* | 50-2.94 (2.99–2.94) |
| Unique reflections* | 54,189 (2664) |
| Rsym (%)* | 9.3 (>100) |
| I/σ(I)* | 23.2 (1.3) |
| Completeness (%)* | 100.0 (100.0) |
| Redundancy* | 8.1 (7.5) |
| Temperature (K) | 100 |
| Mosaicity (°) | 0.45–0.6 |
| Refinement | |
| Resolution (Å) | 48.7-2.94 |
| No. reflections | 53,775 |
| Rwork/Rfree | 0.207/0.245 |
| No. atoms/B-factors (Å2) | |
| CDK9, molecule 1, 2, 3 | 2558 (111.9), 2533 (116.3), 2558 (121.6) |
| Cyclin T1, molecule 1, 2, 3 | 2003 (121.3), 2024 (123.1), 2001 (118.5) |
| AFF4, molecule 1, 2, 3 | 248 (156.3), 421 (161.1), 243 (160.3) |
| Water | 19 (90.1) |
| Root mean square deviations | |
| Bond lengths (Å) | 0.004 |
| Bond angles (°) | 0.666 |
| Ramachandran plot† | |
| Favored (%) | 94.7 |
| Allowed (%) | 4.48 |
| Disallowed (%) | 0.78 |
| Protein Data Bank ID | 4IMY |
-
*
Values in parentheses are for the highest resolution shell.
-
†
Values from MOLPROBITY.
Binding affinities of AFF4 segments
| Direct binding | Competition assay | |||
| AFF432–67 | AFF432–67 | AFF42–73 | AFF42–363 | |
| CycT1 | 104 ± 17 nM | 102 ± 10 nM | 130 ± 18 nM | 115 ± 15 nM |
| P-TEFb | 36 ± 6 nM | 36 ± 4 nM | 10 ± 1 nM | 7 ± 1 nM |
| Tat-P-TEFb | 8.8 ± 0.8 nM | 4.5 ± 0.6 nM | 0.85 ± 0.15 nM | 0.6 ± 0.1 nM |
-
Dissociation constants measured by direct binding of fluorescein-labeled AFF432–67 and by competition with unlabeled AFF4 segments. The increased affinity of AFF4 for P-TEFb compared to CycT1 may be due to structural changes in the cyclin subunit or additional interactions with the CDK9 kinase subunit. The similar affinities of AFF42–73 and AFF42–363 for all the cyclin-containing species suggest that AFF42–73 encompasses the binding sites for P-TEFb and Tat-P-TEFb.
Dissociation constants of AFF432–67 for Cyclin T1 mutants
| CycT1 variant | Kd (nM) |
| Wild-type | 104 ± 17 |
| Y175A | 228 ± 18 |
| E211A | 356 ± 29 |
| D169A | 438 ± 41 |
| F176A | 645 ± 58 |
| R165A | 1592 ± 171 |
| W210A | 2190 ± 246 |
| W207A | 6050 ± 871 |





