Cytoplasmic dynein crosslinks and slides anti-parallel microtubules using its two motor domains
Figures
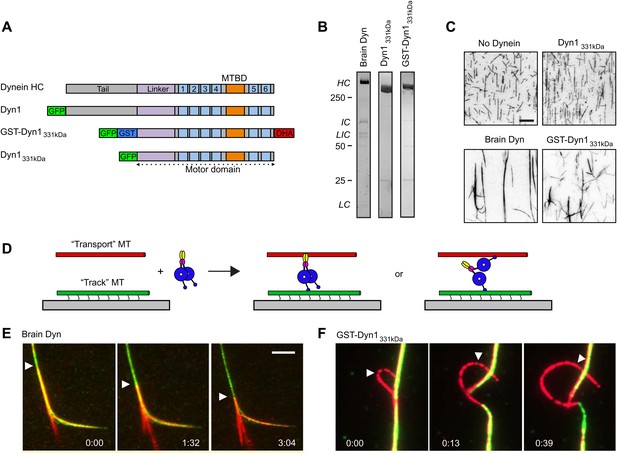
Dynein crosslinks and slides MTs in bundles.
(A) Schematic overview of the dynein constructs used in this study. The N-terminal tail is shown in gray, the linker in purple, the six numbered AAA+ domains are in light blue and the stalk and MT binding domain are depicted in orange. GFP and GST tags are shown in green and blue, respectively. The Halo tag (DHA, Promega) is shown in red. (B) Coomassie brilliant blue stained gels showing purified dynein constructs used in this study. The associated subunits of the brain cytoplasmic dynein complex are labeled; HC–heavy chain, IC–intermediate chain, LIC–light intermediate chain, LC–light chain. Recombinant yeast dynein constructs do not contain associated subunits. Molecular weight markers are indicated. (C) MTs incubated in the absence or presence of dynein are visualized by attachment to a streptavidin-coated coverslip via a biotin tag. Brain dynein and GST-Dyn1331kDa crosslink MTs into large bundles, while the dynein monomer, Dyn1331kDa does not. Scale bar, 10 µm. (D) Cartoon depicting two different mechanisms by which dynein could crosslink MTs, either using its two motor domains or through the tail domain. Alexa-568 and Alexa-488 labeled MTs are crosslinked by dynein. The green MTs are attached to the coverslip through a biotin-streptavidin linkage and perfusion of 1 mM ATP induces sliding between the MTs. (E and F) Example of rat (E) and GST-Dyn1331kDa (F) dynein-driven sliding of red-labeled MTs within the bundle after 1 mM ATP addition. Arrowhead tracks the sliding MT within the bundle. The time relative to the start is noted in min:s at the bottom of each image. Scale bar, 5 µm.
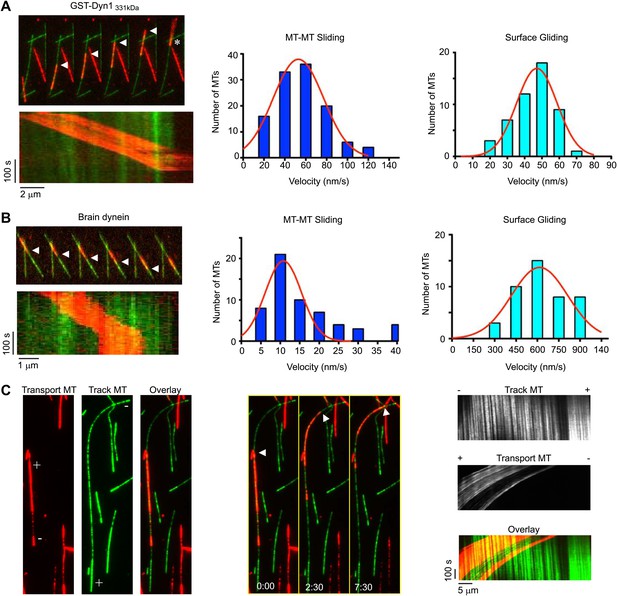
Dynein crosslinks and slides single MT overlaps.
(A) Example of single MT-MT sliding driven by GST-Dyn1331kDa. Successive frames, separated by 52 s, from the video and corresponding kymograph show the sliding. The transport MT is captured and aligned onto the track MT by GST-Dyn1331kDa. Arrowheads follow the transport MT as it slides along the track MT. Asterisk marks where the transport MT remains attached at the end of the track MT. Right, histograms of the MT-MT sliding and surface gliding velocities driven by GST-Dyn1331kDa with Gaussian fitting. (B) Example of rat brain dynein driven sliding in a single MT-MT overlap. Successive frames, separated by 46 s from the video are shown with corresponding kymograph below. The transport MT pauses before reaching the end of the track MT, which was frequently observed for rat dynein-driven movement. Velocity histograms for rat dynein driven MT-MT sliding and surface gliding are shown to the right with Gaussian fitting. (C) Polarity-marked MTs with long, brightly labeled plus-ends were used to determine the orientation of MT-MT sliding. The plus- and minus-ends of both MTs are indicated. Arrowhead shows the red transport MT slides with its minus-end away from the plus-end of the track MT, indicating that sliding is anti-parallel. Kymograph analysis of the anti-parallel sliding event is shown to the right.
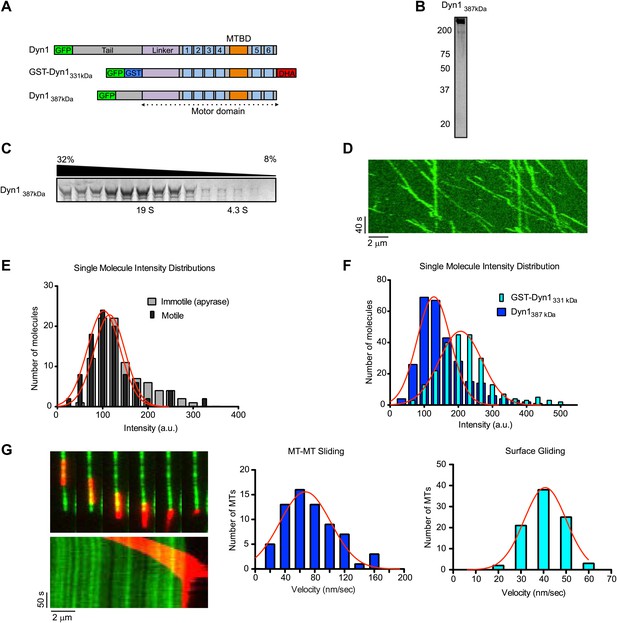
Natively dimerized dynein can drive MT-MT sliding in the absence of associated factors.
(A) Schematic overview of the Dyn1387kD construct compared to Dyn1 and GST-Dyn331kDa, with labels as in Figure 1A. (B) Gel-filtered Dyn1387kD was run on a denaturing gel and proteins were visualized by silver staining. Note the lack of detectable bands at the lower molecular weights expected for dynein accessory subunits. (C) Coomassie blue stained gel of sucrose gradient fractions showing sedimentation behavior of Dyn1387kD. Position of 19S and 4.3S standards and sucrose concentrations are indicated. (D) Kymograph of single Dyn1387kD molecule motility along a MT immobilized on the glass surface. (E) Cy5-labeled MTs were surface-immobilized in flow chambers. Dyn1387kD was added either in the presence of ATP or apyrase. Fluorescence intensities of single motors was determined either of all MT-bound molecules in the presence of apyrase (light gray bars), or of the motile motors in the presence of ATP (dark gray bars). (F) Flow chambers were prepared as in (E), but MTs were incubated with either Dyn1387kD or GST-Dyn1331kD for 5 min in the presence of apyrase. Intensities of GFP spots were quantified after background subtraction. (G) Example of Dyn1387kD driven sliding in a single MT-MT overlap. Successive frames, separated by 34 s from the video are shown with corresponding kymograph below. Velocity histograms for MT-MT sliding and surface gliding are shown to the right with Gaussian fitting.
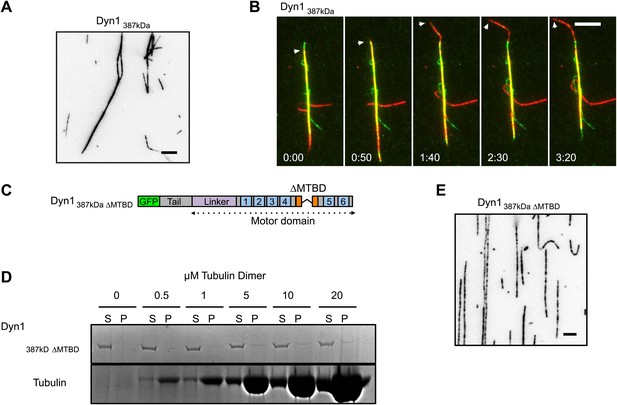
Further characterization of GFP-Dyn1387kD.
(A) Dyn1387kD induces MT bundling in the absence of ATP. Scale bar is 5 μm. (B) Successive frames from Video 5 showing sliding of red MTs in the presence of ATP within a red- and green-labeled MT bundle induced by Dyn1387kD. Scale bar, 5 μm. (C) Schematic showing the construct Dyn1387kD ΔMTBD in which the canonical MTBD has been removed. (D) MT co-pelleting assay demonstrates that GFP-Dyn1387kD ΔMTBD does not specifically bind to MTs. Supernatents (S) and MT pellets (P) are shown at various concentrations of MTs. A small fraction of Dyn1387kD ΔMTBD (≤10%) is found in the pellet at each concentration of tubulin tested. The amount of dynein in the pellet remains constant with increasing tubulin concentration, and is thus assumed to be non-specific binding. (E) Dyn1387kD ΔMTBD does not induce MT bundling in the absence of ATP.
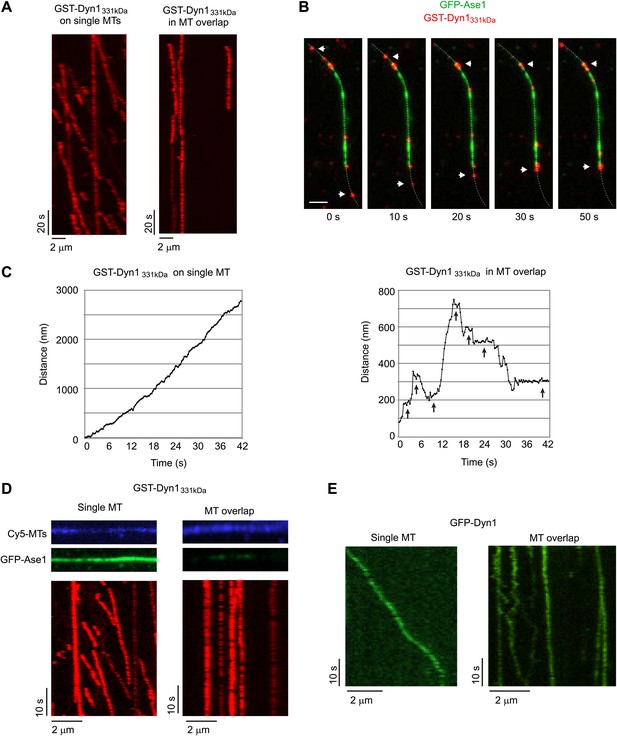
Dynein can bind two MTs simultaneously.
(A–E) Biotinylated Cy5-MTs were incubated with 2 nM GFP-Ase1 or with buffer for 5 min. MTs were then bound to glass slides using surface-bound streptavidin. After washing unbound MTs, TMR-labeled GST-Dyn1331kDa (A–D) or GFP-tagged Dyn1 (E) was introduced into the flow cell. (A) Kymographs of TMR-GST-Dyn1331kDa on single MTs (left panel) and on anti-parallel MT overlaps (right panel). (B) Images from a time series in which individual TMR-GST-Dyn1331kDa molecules can be observed walking into an overlap (visualized by GFP-Ase1) and halting their unidirectional movement. Dotted line indicates the MT. Arrowheads indicate motors that walk from a single MT into an MT overlap. Scale bar is 2 μm. (C) High resolution tracking of individual motors on either single MTs (left) or in an anti-parallel MT overlap (right). Arrows indicate long pauses in motility. Tracking precision was ∼10 nm. (D) Slides were prepared as in (A), but 100 nM of GFP-Ase1 was added to single MTs together with TMR-labeled GST-Dyn1331kDa. GST-Dyn1331kDa shows unidirectional processive movement on Ase1-coated single MTs, but not in Ase1-generated MT overlaps. (E) Slides were prepared as in (A), but full-length yeast Dyn1 was used. Kymographs show dynein moving on a single MT (left) or in a MT overlap created by Ase1 (right).
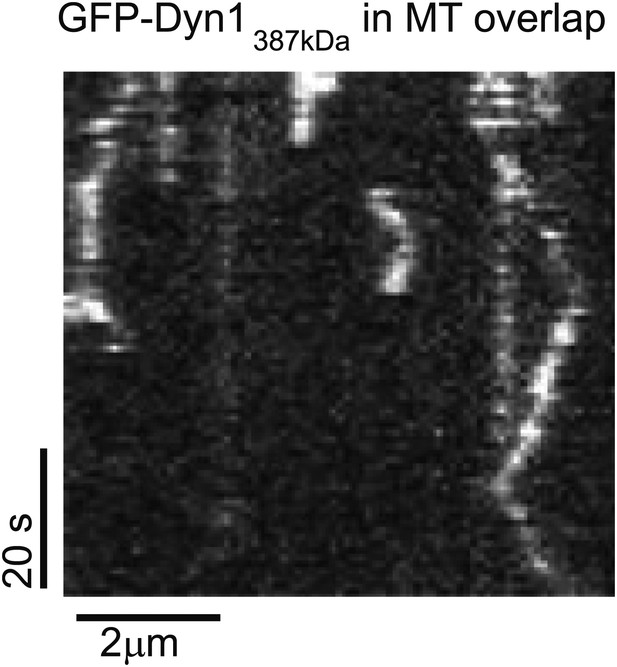
Analysis of single Dyn1387kD molecules in MT overlaps.
Biotinylated Cy5-MTs were incubated with 2 nM GFP-Ase1 for 5 min. MTs were then bound to glass and GFP-Dyn1387kDa was introduced into the flow cell. A kymograph of GFP-Dyn1387kDa within the MT overlap is shown.
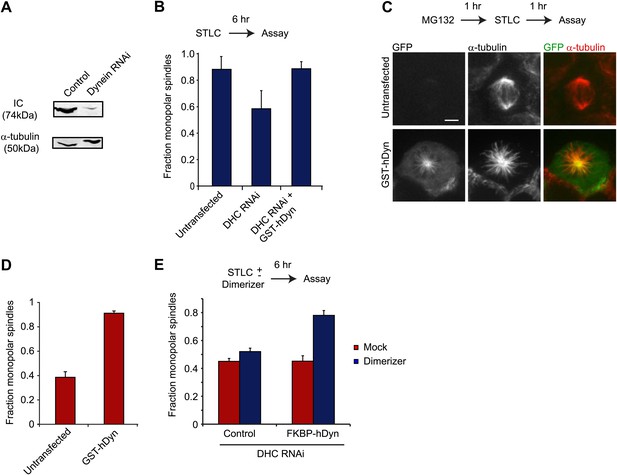
Dynein’s motor domains are sufficient to produce an inward force in the spindle.
(A) HEK293 cells were either mock transfected or transfected with siRNA targeting the dynein heavy chain. After 24 hr cells were re-transfected with siRNA. 72 hr after initial transfection, cells were harvested and the level of dynein expression was determined by western blot. Note that the blot was probed for IC, which is co-depleted when dynein heavy chain is depleted. (B) Cells were either mock transfected or transfected with siRNA targeting the N-terminus of dynein. After 24 hr, cells were washed and transfected with GFP-tagged GST-hDyn. 48 hr after the first transfection, cells were re-transfected with dynein siRNA. 68 hr after the first transfection, cells were treated with 1 µM STLC for 6 hr and were then fixed and stained for α-tubulin. The fraction of mitotic cells with monopolar spindles was then scored. (C and D) Cells were transfected with GFP-tagged GST-hDyn. After 24 hr, cells were treated with MG132 for 1 hr and subsequently with 20 µM STLC for 1 hr where indicated. Cells were then fixed and stained for α-tubulin and the percentage of mitotic cells with monopolar spindles was scored. (C) shows representative images and (D) shows the quantification. (E) Cells were treated as in (C), except GFP-tagged FKBP-hDyn was transfected instead of GST-hDyn. In addition, 200 nM AP20187 was added together with STLC where indicated. Scale bars, 5 µm. Error bars represent standard deviations. All graphs are averages of three independent experiments with 40–120 cells scored per experiment.
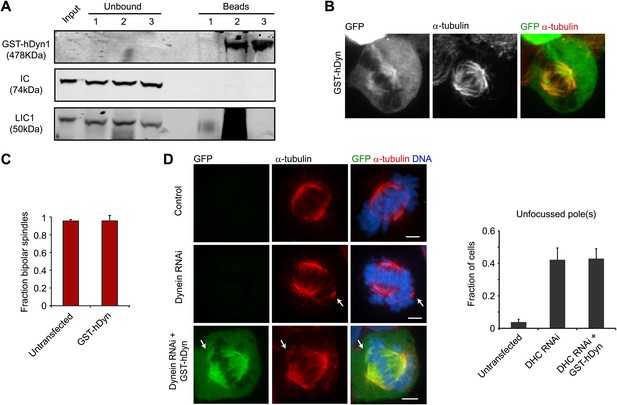
Analysis of GST-hDyn activity in vivo.
(A) HEK293 cells were transfected with GST-hDyn. 24 hr after transfection cells were lysed and pulldowns were performed with either beads alone (1), an HA antibody (2) or a GFP antibody (3) to isolate the GST-hDyn protein. Input, bound and unbound fractions were probed for HA, IC or LIC1. Note the smear in lane 2 is due to antibody heavy chain cross-reactivity. (B) HEK293 cells were transfected with GFP-tagged GST-hDyn. Cells were fixed and stained for α-tubulin. GST-hDyn is observed in the cytoplasm and on the spindle. (C) Cells were transfected with GFP-tagged GST-hDyn. After 24 hr, cells were treated with MG132 for 1 hr. Cells were then fixed and stained for α-tubulin and the percentage of mitotic cells with bipolar spindles was scored. (D) Cells were either mock transfected or transfected with siRNA targeting the N-terminus of dynein. After 24 hr, cells were washed and transfected with GFP-tagged GST-hDyn. 48 hr after the first transfection, cells were re-transfected with dynein siRNA. 68 hr after the first transfection, cells were fixed. Cells were stained for α-tubulin and the degree of spindle pole focusing was determined. Left panel shows representative images. Arrows indicate additional pole. Right panel shows quantification. Scale bars, 5 µm. Error bars represent standard deviations. All graphs are averages of three independent experiments with 30–120 cells scored per experiment.
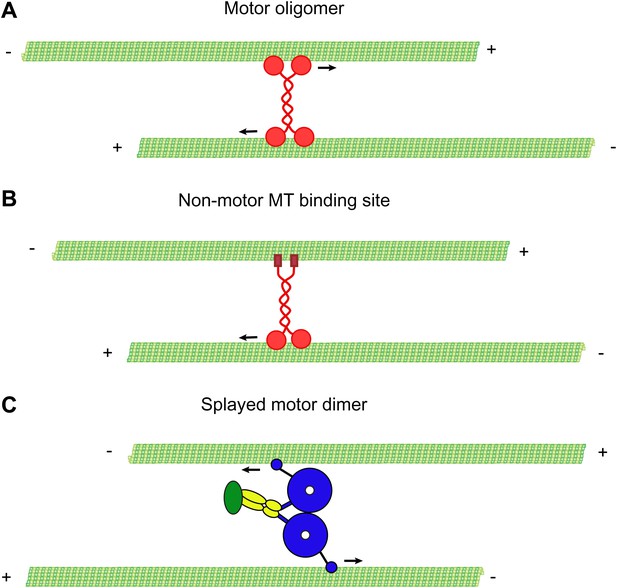
Mechanisms of MT-MT sliding.
(A) Kinesin-5 forms tetrameric molecules, allowing them to crosslink and slide MTs using motor domains at opposite ends of the tetramer. (B) Kinesin-1 and kinesin-14 utilize a secondary, non-motor, MT binding site located in their tail domains to crosslink and slide MTs. (C) Our data suggest that cytoplasmic dynein utilizes a novel mechanism for crosslinking and sliding MTs. The motor domains of a single dynein dimer splay apart and bind to separate MTs. The motor domains then move on opposite MTs, causing sliding of anti-parallel MTs.
Videos
Rat brain cytoplasmic dynein bundles and slides MTs.
Red-labeled MTs are slid and extruded from a bundle of red- and green-labled MT bundles by rat brain cytoplasmic dynein upon addition of ATP. Total time of the video is 551 s. Playback is 30 fps.
GST-Dyn1331kDa slides MTs within bundles.
Red-labeled MTs are extruded from a bundle of red- and green-labeled MTs by GST-Dyn1331kDa upon addition of ATP, indicating the dynein motor domains alone are sufficient for this activity. Total time of the video is 84 s. Playback is 5 fps.
Rat brain cytoplasmic dynein slides single MT overlaps.
A red-labeled ‘transport’ MT is slid over a green-labeled ‘track’ MT by rat brain cytoplasmic dynein. The transport MT pauses before reaching the end of the track MT. Total time of the video is 271 s. Playback is 30 fps.
GST-Dyn1331kDa slides single MT overlaps.
A red-labeled ‘transport’ MT is slid over a green-labeled ‘track’ MT by GST-Dyn1331kDa, indicating that the dynein motor domains alone are sufficient for this activity. The transport MT slides to the end of the track MT and swivels around a nodal attachment point. Total time of the video is 306 s. Playback is 30 fps.
Dyn1387kDa slides MTs within bundles.
Red-labeled MTs are extruded from a bundle of red- and green-labeled MTs by Dyn1387kDa upon addition of ATP. Total time of the video is 250 s. Playback is 12 fps.
Dyn1387kDa slides single MT overlaps.
A red-labeled ‘transport’ MT is slid over a green-labeled ‘track’ MT by Dyn1387kDa. Total time of the video is 198 s. Playback is 12 fps.
Observation of single Dyn1387kDa molecules in sliding MT-MT overlaps.
Single GFP-labeled Dyn1387kDa molecules visualized during a MT-MT sliding event show homogenous fluorescence, further ruling out protein aggregation as a cause of the sliding behavior. Track MT is blue, transport MT is red. Total time of the video is 120 s. Playback is 6 fps.





