The ER–Golgi intermediate compartment is a key membrane source for the LC3 lipidation step of autophagosome biogenesis
Figures
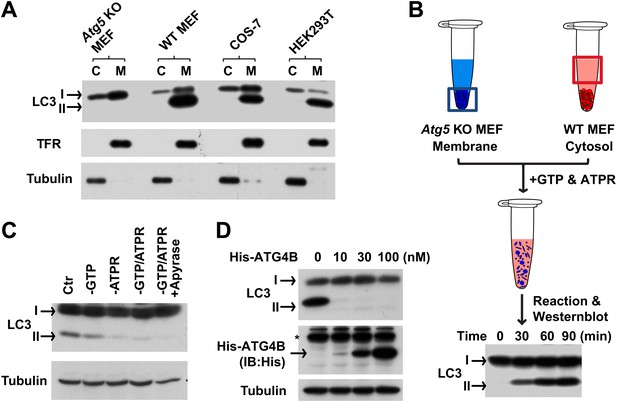
In vitro reconstitution of endogenous LC3 lipidation.
(A) The distribution of LC3-I and LC3-II between the cytosol (C) and membrane (M) fractions from indicated cells. Cytosol and membranes from indicated cells were separated and evaluated by immunoblot (IB) with indicated antibodies. TFR, transferrin receptor (B) cell-free reconstitution of LC3 lipidation. Membranes from Atg5 knockout (KO) MEFs were incubated with cytosol from wild type (WT) cells plus GTP and an ATP regeneration system (ATPR) for the indicated times. Then SDS-PAGE and immunoblot were performed to detect the generation of lipidated LC3 (LC3-II). (C) ATP dependence of in vitro LC3 lipidation. Reactions similar to (B) were performed in the absence or presence of indicated reagents followed by SDS-PAGE and immunoblot. (D) Delipidation of LC3 by ATG4B. A reaction similar to (B) was performed and the 16,000×g membranes were sedimented and solubilized with 1% TritonX-100. The indicated concentrations of ATG4B were incubated with the samples for 30 min followed by SDS-PAGE and immunoblot. Asterisk, non-specific band.
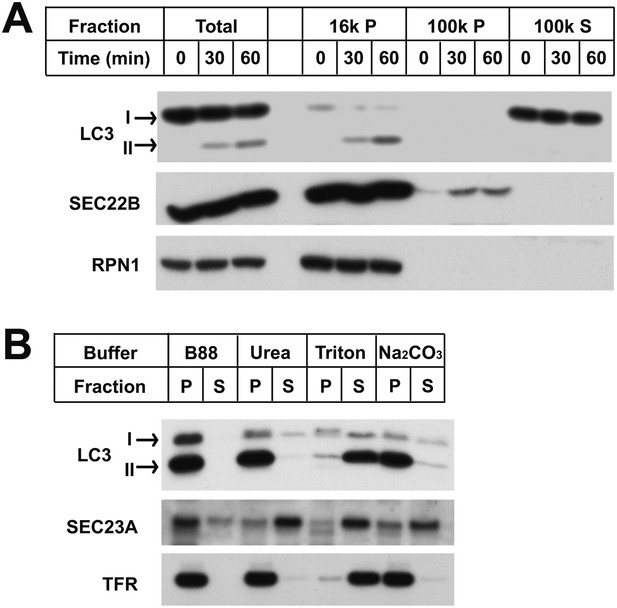
Characterization of the in vitro-lipidated LC3.
(A) LC3-II distributes in the 16,000×g membrane pellet fraction. Reactions similar to those of Figure 1B were performed. After the indicated times, the post-reaction mixtures were centrifuged at 16,000×g for 10 min. The pellet fractions (16k P) were collected and the supernatant fractions were further centrifuged at 100,000×g yielding the pellet (100k P) and supernatant (100k S) fractions. SDS-PAGE and immunoblot were performed with indicated antibodies. RPN1, Ribophorin 1 (B) LC3-II is tightly anchored to membranes. A reaction similar to that in (A) was performed and 16,000×g membranes were collected. The pellet was resuspended, divided into aliquots and incubated with indicated reagents followed by another 16,000×g centrifugation to separate into pellet (P) and supernatant (S) fractions, which were examined by SDS-PAGE and immunoblot. TFR, transferrin receptor.
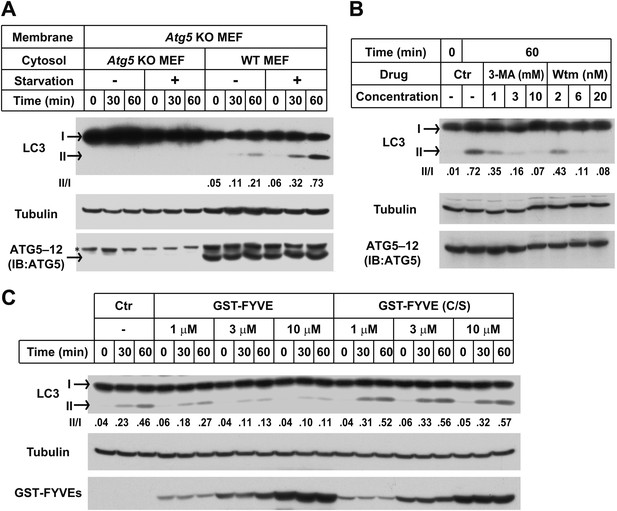
The in vitro lipidation of LC3 is regulated by ATG5, starvation and PI3K.
(A) Starvation-promoted and ATG5-dependent lipidation of LC3. Indicated cells were either untreated or starved for 30 min. The in vitro lipidation reaction with the indicated combination of cytosols and membranes was performed. The formation of LC3-II was analyzed by SDS-PAGE and immunoblot. Asterisk, non-specific band (B) PI3K inhibitors 3-methyladenine (3-MA) and wortmannin (Wtm) inhibit LC3 lipidation. The in vitro lipidation reaction, with cytosol from starved WT MEFs and membrane from Atg5 KO MEFs, was performed in the absence or presence of the indicated concentrations of 3-MA and wortmannin for 60 min. LC3 lipidation was analyzed by SDS-PAGE and immunblot. (C) PI3P dependence of in vitro LC3 lipidation. The in vitro lipidation reaction similar to (B) was performed in the absence or presence of increasing concentrations of GST-FYVE or FYVE (C/S) proteins for the indicated times. SDS-PAGE and immunoblot were performed to analyze the level of LC3-II. Quantification of lipidation activity was shown as the ratio of LC3-II to LC3-I (II/I).
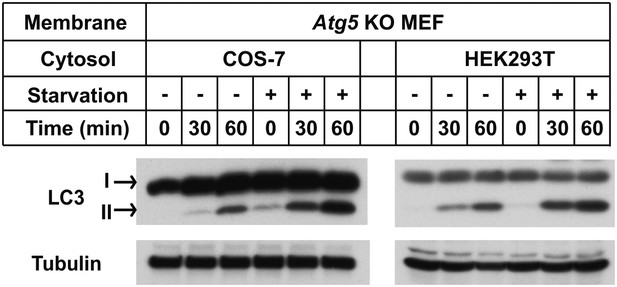
Starvation-promoted lipidation of LC3 by COS-7 or HEK293T cytosol.
COS-7, HEK293T and Atg5 KO MEF cells were either untreated or starved for 60 min. The in vitro lipidation reaction with indicated combination of cytosols and membranes was performed followed by SDS-PAGE and immunoblot to examine the formation of LC3-II.
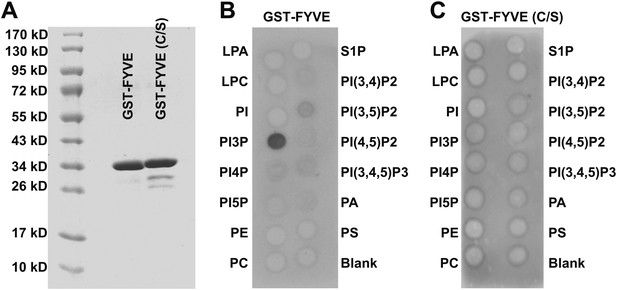
Purification and verification of GST-FYVEs.
(A) Purification of GST-fusion PI3P binding FYVE domains and mutants (C/S). (B) PIP Strip blot with 10 µg/ml GST-FYVE. (C) PIP Strip blot with 10 µg/ml GST-FYVE (C/S).
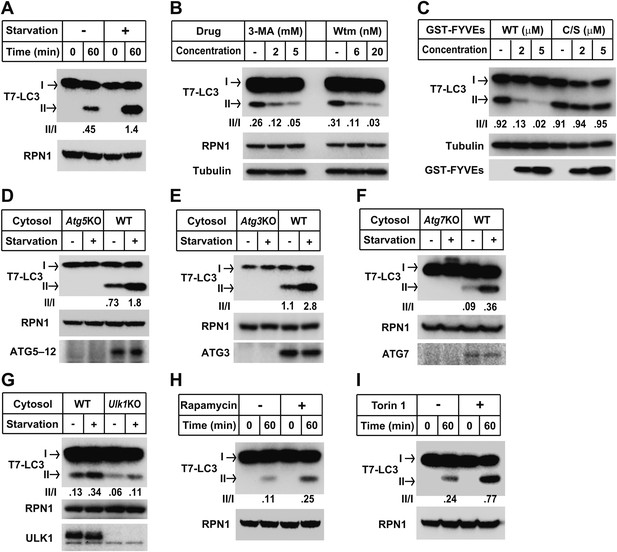
Recapitulation of the major regulatory pathways for autophagy by in vitro lipidation of T7-LC3.
(A) Starvation-induced lipidation of T7-LC3. HEK293T and Atg5 KO MEF cells were either untreated or starved for 90 min. The in vitro lipidation reaction was performed by incubating T7-LC3 with HEK293T cytosols and Atg5 KO MEF membranes with indicated treatments for the indicated times followed by SDS-PAGE and immunoblot. (B) 3-methyladenine and wortmannin inhibit T7-LC3 lipidation. The in vitro lipidation reaction was performed by incubating T7-LC3 with cytosol from starved HEK293T and Atg5 KO MEF membranes in the absence or presence of the indicated drugs for 60 min followed by SDS-PAGE and immunoblot. (C) PI3P dependence of in vitro T7-LC3 lipidation. In vitro lipidation reactions similar to (B) were performed in the absence or presence of the indicated concentrations of GST-FYVEs for 60 min followed by SDS-PAGE and immunoblot. (D) Dependence on ATG5 for T7-LC3 lipidation. The in vitro lipidation reaction was performed by incubating T7-LC3 with starved cytosols as indicated and Atg5 KO MEF membranes for 60 min followed by SDS-PAGE and immunoblot to analyze LC3-II in the membrane fraction. (E) Dependence on ATG3 for T7-LC3 lipidation. A similar experiment was performed using cytosols from Atg3 KO and WT MEFs, and membrane from Atg3 KO MEFs. (F) Dependence on ATG7 for T7-LC3 lipidation. A similar experiment was performed using cytosols from Atg7 KO and WT MEFs, and membrane from Atg7 KO MEFs. (G) Dependence on ULK1 for starvation-induced T7-LC3 lipidation. The in vitro lipidation reaction was performed by incubating T7-LC3 with untreated or starved cytosols as indicated and Ulk1 KO MEF membranes for 60 min followed by SDS-PAGE and immunoblot of the membrane fraction. (H) Rapamycin-induced lipidation of T7-LC3. Cells were treated with 1 µM rapamycin or a control solution for 2 hr and cytosol was incubated with membranes as in (A). (I) Torin 1-induced lipidation of T7-LC3. Cells were treated with 200 nM Torin 1 or a control solution for 90 min and incubated with membranes as above. Quantification of lipidation activity was shown as the ratio of LC3-II to LC3-I (II/I).
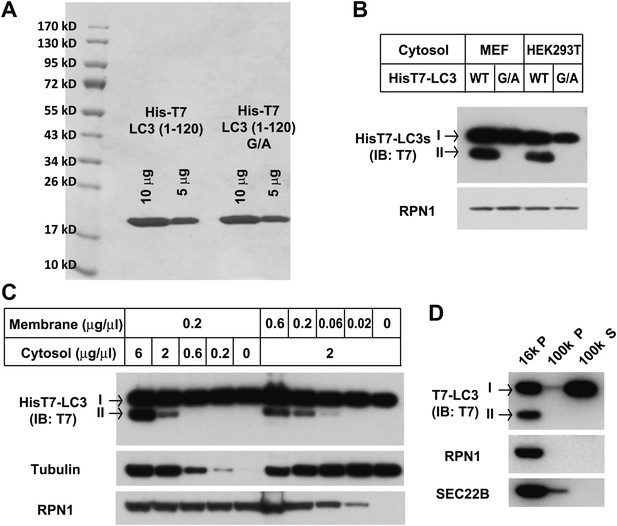
Purification of the T7-tagged LC3 and characterization of the lipidation.
(A) Purification of HisT7-LC3 and the G/A mutant. (B) Lipidation of HisT7-LC3. The in vitro lipidation reaction was performed by incubating HisT7-LC3 or G/A mutant with indicated cytosols and Atg5 KO MEF membranes for 60 min followed by SDS-PAGE and immunoblot. (C) Cytosol and membrane dependence of HisT7-LC3 lipidation. The in vitro lipidation reaction was performed by incubating HisT7-LC3 with increasing concentrations of cytosol or membrane for 60 min followed by SDS-PAGE and immunoblot. (D) T7-LC3-II distributes in the 16,000×g membrane pellet fraction. T7-LC3 protein was generated by thrombin digestion of the HisT7-LC3 protein to remove the N-terminal His tag. The in vitro lipidation reaction was performed by incubating T7-LC3 with cytosol from starved HEK293T cells and Atg5 KO MEF membranes for 60 min. A centrifugation procedure similar to the experiment in Figure 1—figure supplement 1A was employed and the reaction products were evaluated by SDS-PAGE and immunoblot.
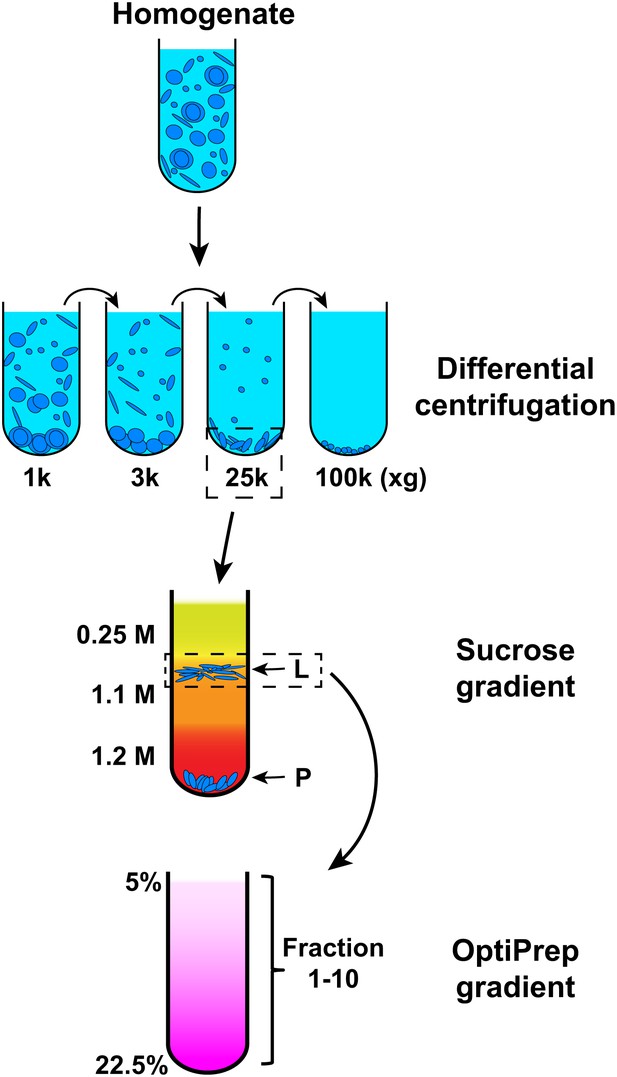
Membrane fractionation scheme.
Briefly, Atg5 KO MEFs were homogenized and the lysates were subjected to differential centrifugations with indicated g forces. The ability of each fraction to trigger T7-LC3 lipidation was examined. The 25,000×g (25k) pellet, which had the most activity, was selected and a sucrose gradient ultracentrifugation was performed to separate the 25k pellet to L (light) and P (pellet) fractions. The L fraction, which contained the majority of the activity to promote T7-LC3 lipidation, was further resolved on an OptiPrep gradient after which ten fractions from the top were collected and the lipidation activity was examined in each.
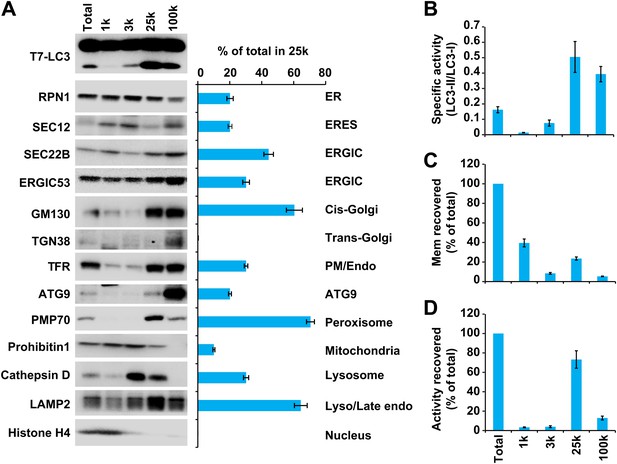
Separation of the total membrane by differential centrifugations.
(A–D) A differential centrifugation experiment was performed as depicted in Figure 4. The total PC of each fraction was measured and presented as a percentage of the total membrane (C) and adjusted to a concentration of 0.6 mg/ml. The T7-LC3 lipidation activity of each fraction was tested and immunoblot was performed to examine the generation of lipidated T7-LC3 as well as the distribution of the indicated membrane markers (A). The level of each marker in the 25k pellet fraction was calculated as a percentage of the total membrane (A). The specific activity (the ability of each membrane fraction to induce LC3 lipidation with the equal amount of PC) of each membrane fraction to trigger T7-LC3 lipidation was measured as a ratio of lipidated to unlipidated T7-LC3s (B). The total activity recovered from each fraction was calculated by multiplying the specific activity by the corresponding PC level of each fraction and shown as a percentage of the total membrane (D). Error bars represent standard deviations of at least three experiments. RPN1, Ribophorin1; TFR, Transferrin receptor; Mem, membrane; Endo, endosome.
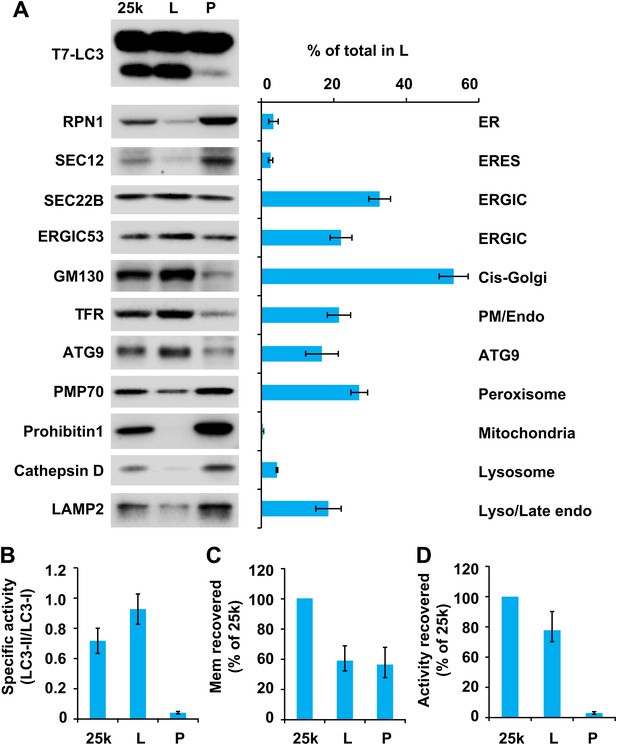
Separation of the 25k pellet fraction by sucrose gradient ultracentrifugation.
(A–D) A sucrose step gradient ultracentrifugation to further separate the 25k pellet fraction was performed as depicted in Figure 4. The total PCs of each fraction were measured and presented as a percentage of the 25k pellet membrane (C) and adjusted to a concentration of 0.6 mg/ml. The T7-LC3 lipidation activities of the L and P fraction were tested and immunoblot was performed as in Figure 5A. The level of each marker in the L fraction was calculated as a percentage of the total membrane (A). The specific activity of each membrane fraction was measured as in Figure 5B. The total activity recovered from each fraction was calculated by multiplying the specific activity by the PC level of each fraction and shown as the percentage of 25k pellet membrane (D). Error bars represent standard deviations of at least three experiments.
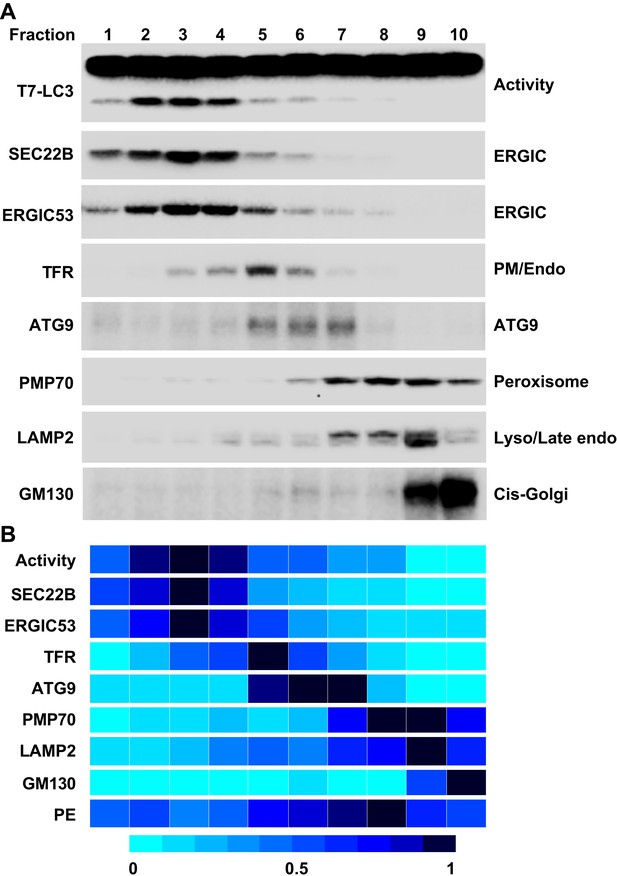
Separation of the L fraction by OptiPrep gradient ultracentrifugation.
(A–B) An OptiPrep gradient ultracentrifugation was used to resolve membranes in the L fraction, as depicted in Figure 4. 10 fractions were collected. The total PCs of each fraction were measured and adjusted to a concentration of 0.6 mg/ml. The T7-LC3 lipidation activities of each fraction were tested and immunoblot was performed as in Figure 5A. The specific activity of each membrane fraction was measured similar to Figure 5. The PE level of each normalized fraction was determined. A heat map showing the relative levels of the specific activity, PE and each of the indicated markers was generated (B). In each group the fraction with the highest value was defined as 1.
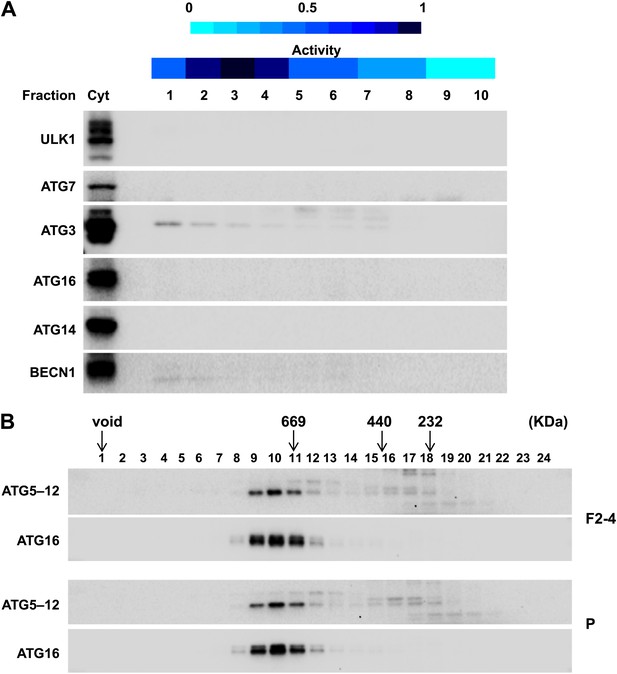
The ERGIC membrane promotes LC3 lipidation without altering cytosolic factors.
(A) The major autophagy factors are cytosolic. Immunoblot of the fractions from Figure 7 and the cytosol (Cyt) used for the in vitro lipidation assay was performed with indicated antibodies. (B) The formation of ATG5–12–16 complex is not altered by ERGIC. Indicated membrane fractions obtained from the membrane fractionation were incubated with cytosol for the LC3 lipidation assay. Membranes were removed by centrifugation and the supernatant fractions were collected for size exclusion chromatography on a Superpose 6 column followed immunoblot analysis with the indicated antibodies. Most of the ATG5 is conjugated with ATG12 in wild-type cells (data not shown). ERGIC, the combination of fractions 2–4 in the OptiPrep gradient fractionation; P, the pellet fraction from the sucrose gradient centrifugation.
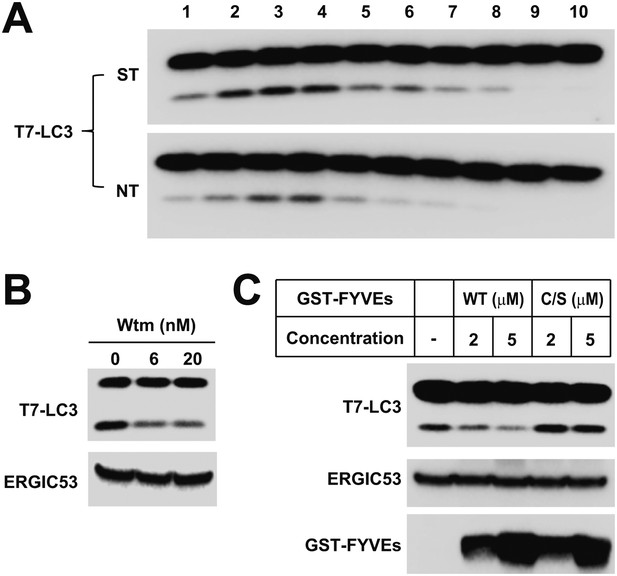
The lipidation activity of the ERGIC-enriched fractions are regulated by starvation and PI3K.
(A) Lipidation of T7-LC3 from the ERGIC-enriched fractions is enhanced by starvation. An in vitro lipidation reaction similar to that in Figure 7 was performed with HEK293T cytosols from starved (ST) and non-starved (NT) cells followed by SDS-PAGE and immunoblot. (B) Wortmannin inhibits T7-LC3 lipidation triggered by the ERGIC-enriched fraction. Fractions 2–4 from Opti-prep gradient were collected and pooled. The in vitro lipidation reaction was performed with increasing concentrations of wortmannin (Wtm) followed by SDS-PAGE and immunoblot. (C) PI3P dependence of T7-LC3 lipidation triggered by the ERGIC-enriched fraction. The ERGIC-enriched membranes were pooled as shown in (B). The in vitro lipidation reaction was performed with indicated concentrations of GST-FYVE and FYVE(C/S) followed by SDS-PAGE and immunoblot.
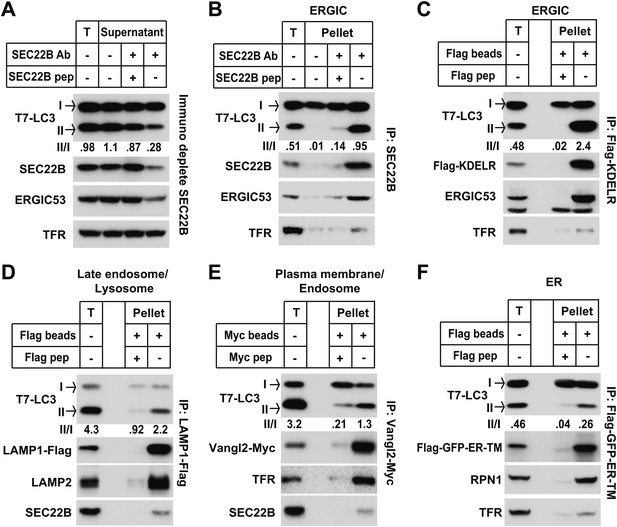
ERGIC directly triggers in vitro LC3 lipidation.
(A) Immunodepletion of ERGIC membrane from L fraction reduces in vitro lipidation activity. The L fraction was prepared as shown in Figures 4 and 5. An immunodepletion experiment with indicated combinations of anti-SEC22B antibody (Ab) and blocking peptide (pep) was performed. The membranes from the flow-through were collected and the in vitro lipidation reaction was performed. Equal amounts of membrane from each group were used for the lipidation reaction. T, total membrane from the L fraction. (B) Enrichment of lipidation activity on the SEC22B-positive membranes. Immunoisolation of SEC22B positive membranes from the L fraction of Atg5 KO MEFs was performed and the in vitro lipidation reaction was conducted with membranes bound to the beads as well as the total membrane (T) from the L fraction. The total membrane used was adjusted to the same amount of the membranes (based on PC content) specifically bound to the beads in the reaction. (C) Enrichment of lipidation activity on KDEL Receptor (KDELR)-positive membranes. Atg5 KO MEFs were transfected with a plasmid encoding the Flag-tagged KDELR protein. 48 hr after transfection, KDELR-positive membranes were immunoisolated with anti-Flag agarose and assayed for lipidation activity as in (B). (D–F) Lipidation activity was not enriched in late endosome/lysosome, plasma membrane/endosome or ER membranes. Atg5 KO MEFs were transfected with plasmids encoding LAMP1-Flag (D), Vangl2-Myc (E) or Flag-GFP-ER-TM (F). 48 hr after transfection, the 25k pellet fractions (for LAMP1-Flag and Flag-GFP-ER-TM) or the L fraction were collected and immunoisolations were performed with anti-Flag agarose or anti-Myc agarose as described above and assayed for lipidation activity. Quantification of lipidation activity was shown as the ratio of LC3-II to LC3-I (II/I).
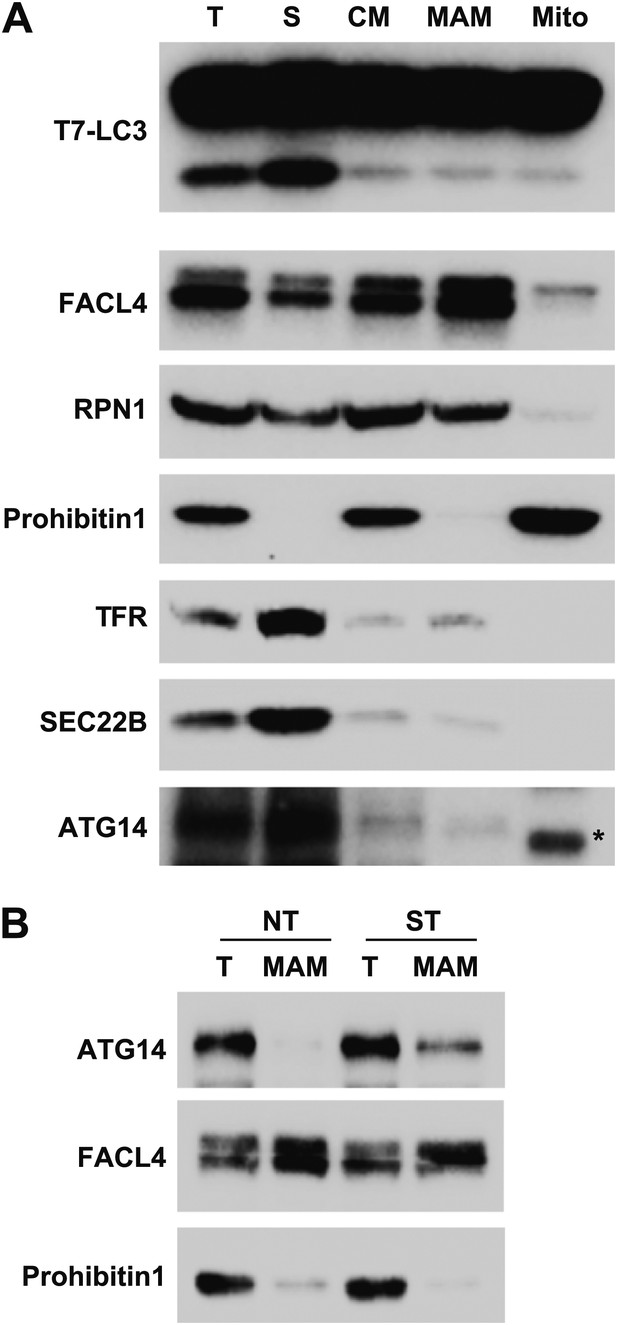
Mitochondrial-associated endoplasmic reticulum membranes (MAM) are not active to trigger in vitro LC3 lipidation.
(A) Indicated membrane fractions were prepared as described by Wieckowski et al. (Wieckowski et al., 2009) and in vitro lipidation was performed as shown in Figures 4–7. T, total membrane; S, supernatant membrane separated from crude mitochondria preparation; CM, crude mitochondria fraction; Mito, mitochondria fraction; MAM, Mitochondrial-associated endoplasmic reticulum membranes, FACL4, Long-chain acyl-CoA synthetase 4. (B) Immunoblot analysis of the MAM fractions purified from non-treated (NT) and starved (ST) cells. Asterisk, non-specific band.
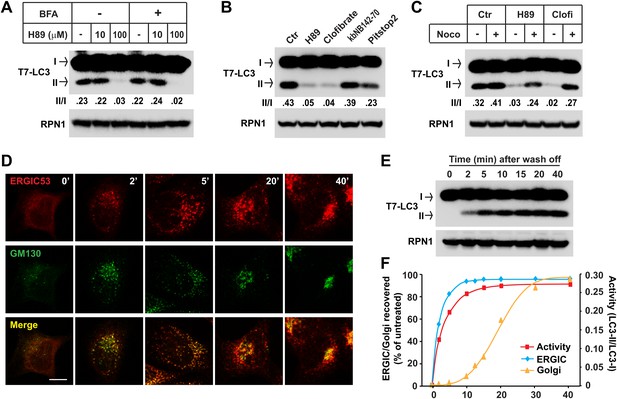
ERGIC is required for in vitro LC3 lipidation.
(A and B) In vivo depletion of ERGIC abolishes the in vitro lipidation of LC3. Atg5 KO MEFs were treated without or with 10 µg/ml Brefeldin A (BFA) for 30 min and then incubated with the indicated concentrations of H89 for 10 min (A). Alternatively, cells were directly treated with the indicated drugs: 100 µM H89, 500 µM clofibrate, 50 µM kbNB142-70 and 50 µM Pitstop2 (B). Total membranes from the cells were collected, the lipidation reaction with T7-LC3 was performed and the products evaluated by SDS-PAGE and immunoblot. Ctr, control (C) Blocking ERGIC disruption preserved the in vitro lipidation of LC3. Atg5 KO MEFs were treated with control, H89 or clofibrate (Clofi) in the absence or presence of nocodazole (Noco). Lipidation reactions with the total membranes from the treated cells were performed. (D–F) The in vitro lipidation of LC3 recovers with restoration of ERGIC. Atg5 KO MEF cells were treated with BFA followed by 100 µM H89. Cells were then washed with fresh medium to remove the drugs and, at indicated intervals, samples were collected for immunofluorescence (D) or total membrane collection for the in vitro lipidation reaction (E). Quantification of the recovery of lipidation activity, ERGIC and Golgi are shown in (F). Bar, 10 µm. Quantification of lipidation activity was shown as the ratio of LC3-II to LC3-I (II/I).
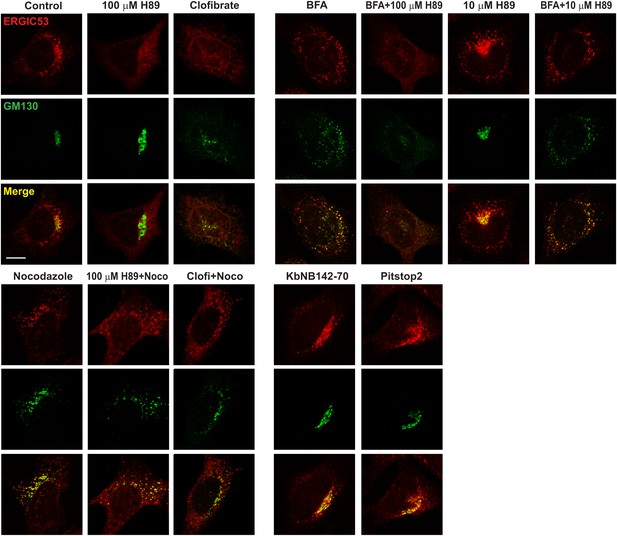
Immunofluorescence showing the effect of indicated drugs on ERGIC and Golgi.
Atg5 KO MEFs were treated with indicated drugs or drug combinations as depicted in Figure 9A,B. Cells were fixed for immunofluorescence with the indicated antibodies. Bar, 10 µm.
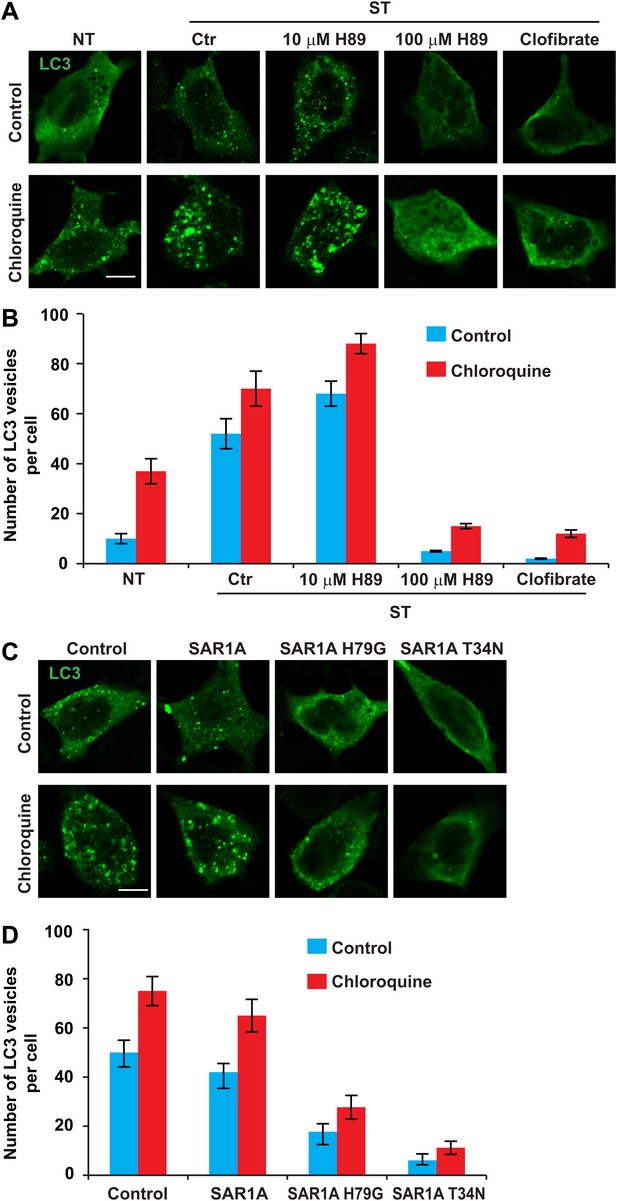
ERGIC is required for starvation-induced LC3 puncta formation.
(A) Drugs that disrupt ERGIC inhibit LC3 puncta formation. MEFs were transfected with plasmids encoding Myc-LC3. After transfection (24 hr), the cells were either non-starved (NT) or starved (ST) in the absence or presence of the indicated drugs followed by immunofluorescence using anti-Myc antibody. Bar, 10 µm. (B) Quantification of the cells shown in (A). Error bars represent standard deviations of three experiments. (C) Genetically disrupting ERGIC inhibits LC3 puncta formation. MEF cells were co-transfected with plasmids encoding Myc-LC3 and the indicated SAR1A variants. After transfection (24 hr), the cells were starved in the absence or presence of chloroquine followed by immunofluorescence using anti-Myc antibody. Bar, 10 µm. (D) Quantification of the cells shown in (C). Error bars represent standard deviations of three experiments.
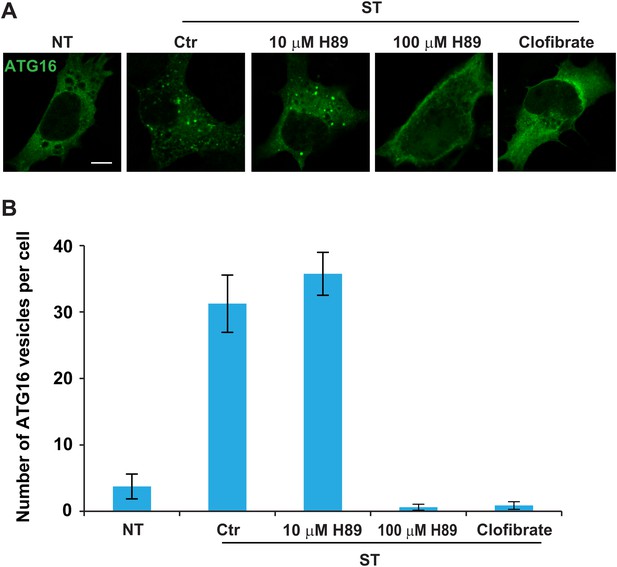
Drugs that disrupt ERGIC inhibit starvation-induced ATG16 puncta formation.
(A) MEFs were transfected with plasmids encoding ATG16-Myc. After transfection (24 hr), the cells were either non-starved (NT) or starved (ST) in the absence or presence of the indicated drugs followed by immunofluorescence using anti-Myc antibody. Bar, 10 µm (B) Quantification of the cells shown in (A). Error bars represent standard deviations of three experiments.
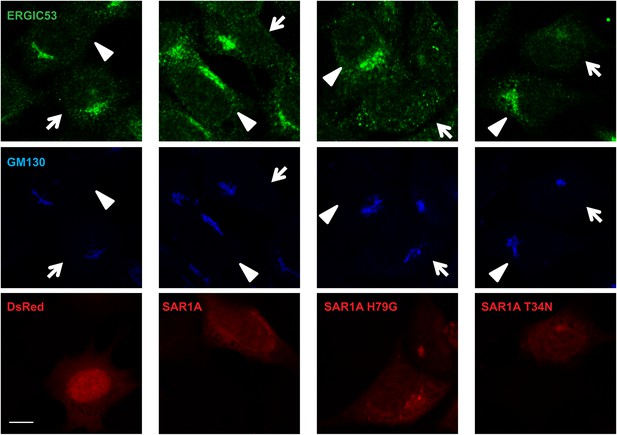
Effects of SAR1A variants on ERGIC.
MEFs were transfected with plasmids encoding the indicated SAR1A-DsRed variants or control DsRed. After transfection (24 hr), immunofluorescence with indicated antibodies was performed. Bar, 10 µm.
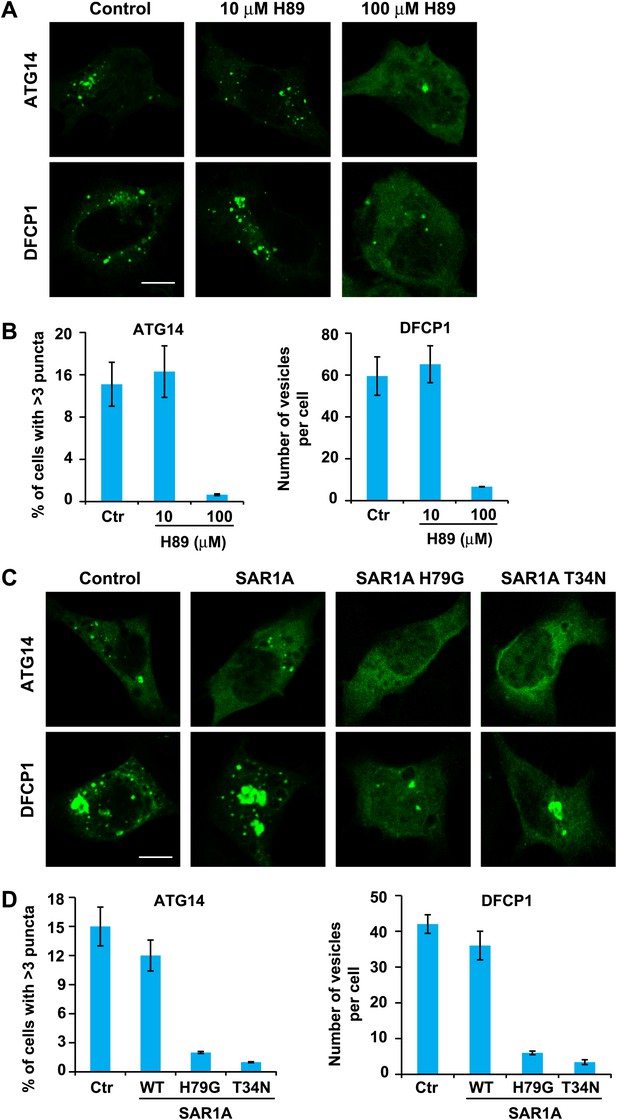
ERGIC is required for the starvation-induced localization of ATG14 and DFCP1 to puncta.
(A) H89 inhibits ATG14 and DFCP1 puncta formation. MEF cells were transfected with plasmids encoding EGFP-tagged ATG14 or DFCP1. After transfection (24 hr), cells were starved in the absence or presence of the indicated concentrations of H89 followed by fixation and direct visualization of the EGFP signal. Bar, 10 µm. (B) Quantification of the cells shown in (A). Error bars represent standard deviations of three experiments. (C) Expression of SAR1A mutants inhibits the formation of puncta that contain ATG14 and DFCP1. MEF cells were co-transfected with plasmids encoding EGFP-tagged ATG14 or DFCP1 and indicated SAR1A-DsRed variants. After transfection (24 hr), cells were starved followed by fixation and direct visualization of EGFP signal. Bar, 10 µm. (D) Quantification of the cells shown in (C). Error bars represent standard deviations of three experiments.
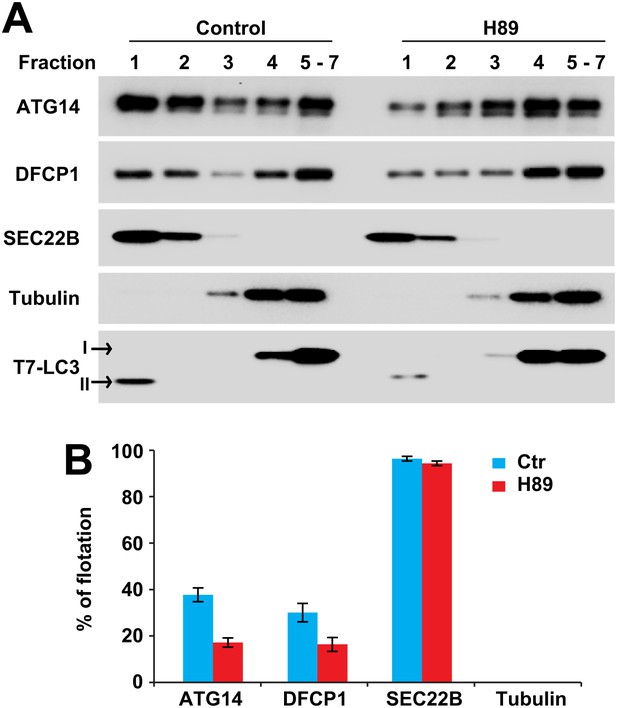
ERGIC is required for membrane recruitment of ATG14 and DFCP1.
(A) Disruption of ERGIC inhibits membrane recruitment of ATG14 and DFCP1. Atg5 KO MEFs were either untreated or treated with H89. Membranes were collected and incubated with cytosol of HEK293T cells expressing ATG14-HA and EGFP-DFCP1. A buoyant density flotation assay was performed followed by immunoblot. (B) Quantification of the floated markers shown in (A). The quantification of samples from the buoyant density gradient was calculated as the ratio of chemiluminescence in the first two fractions to the sum of all fractions. Error bars represent standard deviations of three experiments.
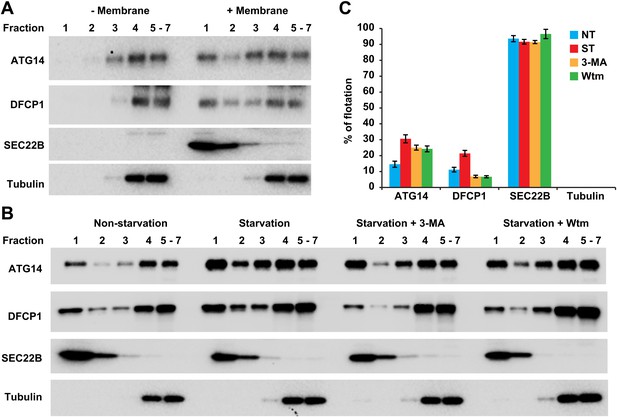
Establishment of the in vitro membrane recruitment assay.
(A) Membrane dependence for the flotation of ATG14 and DFCP1. Cytosol from HEK293T cells expressing ATG14-HA and EGFP-DFCP1 was incubated with or without membrane. A buoyant density gradient flotation assay was performed followed by SDS-PAGE and immunoblot. (B) Starvation and PI3K regulation of the membrane recruitment of ATG14 and DFCP1. Atg5 KO MEF membranes were incubated with the cytosol from either non-starved or starved HEK293T cells expressing ATG14-HA and EGFP-DFCP1 in the presence of indicated drugs. A buoyant density flotation was performed and samples were evaluated by SDS-PAGE and immunoblot. (C) Quantification of the floated markers shown in (B). Error bars represent standard deviations of three experiments.
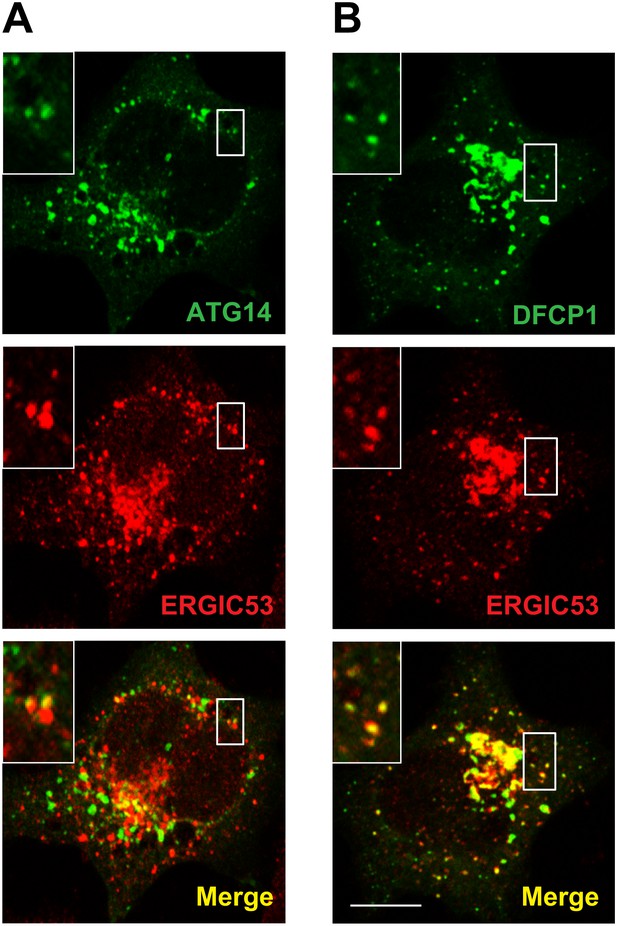
Atg14L and DFCP1 puncta colocalize with ERGIC.
(A and B) MEF cells were transfected with plasmids encoding ATG14-EGFP (A) or EGFP-DFCP1 (B). 24 hr after transfection, the cells were starved for 20 min. Cells were then fixed and visualized by immunofluorescence with anti-ERGIC53 antibody. Insets show the magnified view of the boxed areas. Bar, 10 µm.





