A secreted Ustilago maydis effector promotes virulence by targeting anthocyanin biosynthesis in maize
Figures
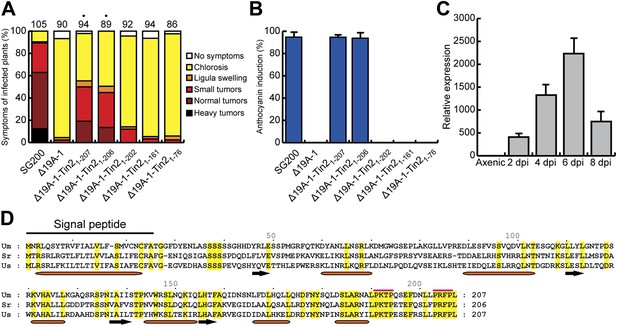
Expression and mapping of functional domains in Tin2.
(A) Biological function of truncated Tin2 proteins. Virulence of SG200Δ19A-1 strains expressing either wild-type or C-terminally truncated Tin2 proteins. Numbers indicate total infected plants. Dots indicate that at least one of the symptoms (ligula swelling, small tumors, normal tumors, heavy tumors) was significantly changed relative to SG200Δ19A-1. (B) The same infected plants as in (A) were scored for anthocyanin pigmentation. Percentage of plants displaying anthocyanin coloration is indicated. Error bars depict standard deviation. (C) Quantification of tin2 gene expression during biotrophic development of U. maydis. Total RNA was extracted from leaves infected with SG200 at 2, 4, 6, and 8 days post infection (dpi), and also from cells grown in axenic culture in YEPSL medium. Transcript levels of tin2 during different growth stages of U. maydis strain SG200 were determined by quantitative real-time PCR. Constitutively expressed U. maydis peptidylprolyl isomerase (ppi) was used for normalization. Three biological replicates were analyzed, error bars depict standard deviation. tin2 expression in budding cells grown in axenic culture was set to 1.0. (D) Amino acid sequence alignment of Tin2 proteins (Um, U. maydis; Sr, S. reilianum; Us, U. scitaminea). Identical amino acids are boxed in yellow. Alpha helices (orange bars) and beta sheets (black arrows) are indicated. Proline-repeat sequences are marked with red lines.
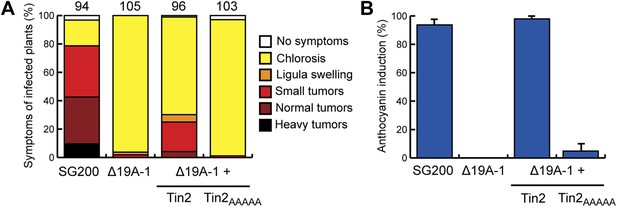
Substitution of the C-terminal 5 amino acids of Tin2 affects effector function.
The C-terminal 5 amino acids (PRFPL) of Tin2 protein were substituted by alanine (AAAAA) and the respective gene (tin2AAAAA) was introduced in single copy in SG200Δ19A-1. (A) Virulence of SG200Δ19A-1 and the derived strains expressing Tin2 or Tin2AAAAA was scored at 12 dpi following the scheme developed by Kämper et al. (2006). The total number of infected plants is indicated above each panel. The expression of Tin2AAAAA did not complement tumor formation. (B) The same infected plants as in (A) were scored for anthocyanin induction. Tin2AAAAA very weakly complemented anthocyanin induction.
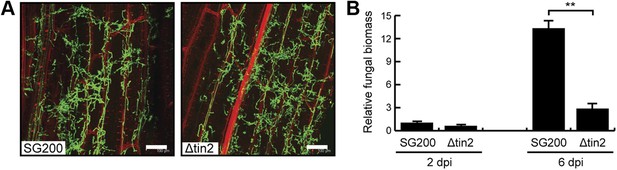
Biotrophic development of SG200 and SG200Δtin2.
(A) Maize seedlings were infected by U. maydis strain SG200 and SG200Δtin2 and observed at 3 days post inoculation by confocal microscopy. Fungal hyphae were visualized by WGA-AF488 staining (green). Plant cell walls were visualized by propidium iodide staining (red). Bar = 100 µm. (B) Quantification of fungal biomass in SG200- and SG200Δtin2-infected tissue. Genomic DNA was extracted from leaf segments infected with SG200 and SG200Δtin2 at 2 and 6 days post inoculation. Quantitative real-time PCR was performed on genomic DNA. Relative fungal biomass was calculated from the U. maydis ppi gene and the Z. mays GAPDH gene, after setting the ratio of SG200 at 2 dpi to 1.0. Error bars indicate the standard deviation of three biological replicates. **p<0.01.
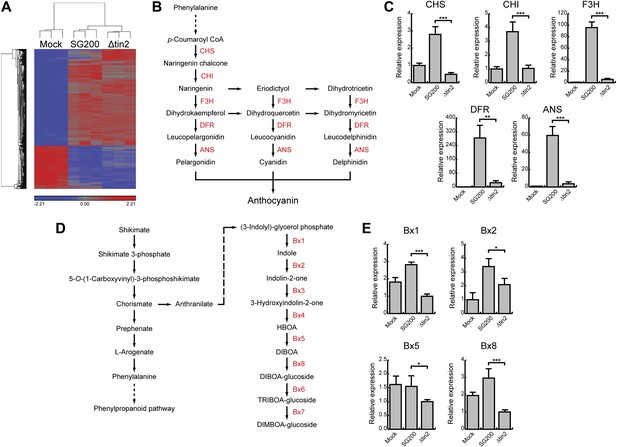
Differentially expressed maize genes in leaves infected with SG200 and SG200Δtin2.
(A) Plant transcriptome analysis of SG200- and SG200Δtin2-infected tissue. Hierarchical clustering was performed by the Partek Genomics Suite version 6.12 to visualize expression of maize genes transcriptionally regulated 4 days after mock inoculation (left) infection by U. maydis strain SG200 (middle) and infection by SG200Δtin2 (right). The X-axis depicts clustering of the microarray samples for each of the three biological replicates per inoculation. The Y-axis shows clustering of the regulated maize transcripts based on similarity of their expression patterns. Red: upregulated genes; blue: downregulated genes. (B) Schematic model of the flavonoid biosynthetic pathway in maize. The enzymes involved in the discrete biosynthetic steps are shown in red. CHS, chalcone synthase; CHI, chalcone isomerase; F3H, flavanone 3-hydroxylase; DFR, dihydroflavonol 4-reductase; ANS, anthocyanin synthase. (C) qPCR based quantification of the expression levels of genes from the flavonoid pathway after infection with indicated U. maydis strains and collecting infected leaf material 6 dpi. **p<0.01, ***p<0.001. (D) Schematic model of the DIMBOA biosynthetic pathway in maize. The enzymes involved in the discrete biosynthetic steps are shown in red. Bx1, indole-3-glycerol phosphate lyase; Bx2, indole monooxygenase; Bx3, indolin-2-one monooxygenase; Bx4, 3-hydroxyindolin-2-one monooxygenase; Bx5, HBOA monooxygenase; Bx8, DIBOA UDP-glucosyltransferase; Bx6, 2-oxoglutarate-dependent dioxygenase; Bx7, TRIBOA-glucoside methyltransferase. (E) qPCR based quantification of the expression levels of genes from the DIMBOA pathway after infection with indicated U. maydis strains and collecting infected leaf material 6 dpi. *p<0.05, ***p<0.001.
-
Figure 3—source data 1
List of upregulated maize genes after SG200 infection (vs SG200Δtin2).
- https://doi.org/10.7554/eLife.01355.007
-
Figure 3—source data 2
List of differentially regulated maize genes after SG200 infection (vs mock).
- https://doi.org/10.7554/eLife.01355.008
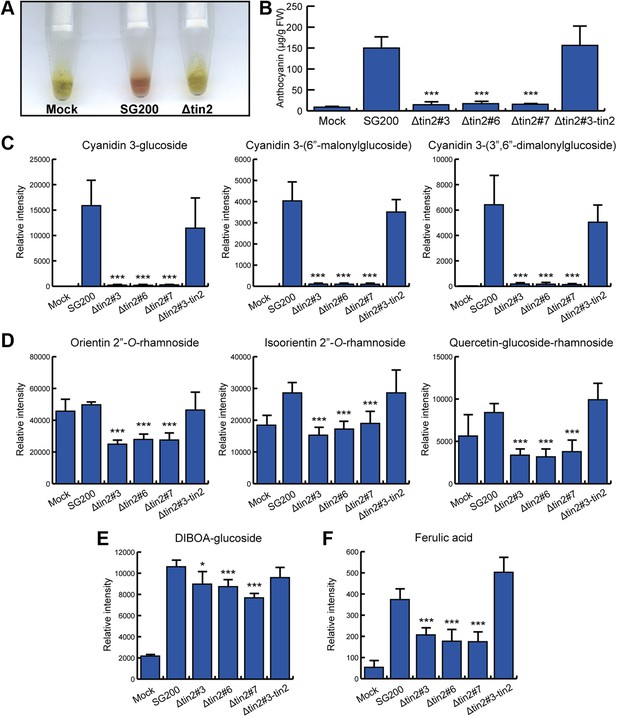
Identification of anthocyanins and other metabolites in infected maize leaves.
(A) Anthocyanin was extracted from leaves syringe-inoculated with H2O (mock), SG200, and SG200Δtin2#3. Two cm long leaf segments located 1 cm below the injection holes were excised, frozen in liquid nitrogen, ground and extracted with 100% ethanol. For each sample, 5 leaf segments were combined. The supernatant from SG200-infected leaves showed red color after acidification with concentrated HCl while the other two samples stayed green. (B–E) Infected leaf segments from mock-inoculated leaves and leaves infected with SG200, three independent SG200Δtin2 mutants and SG200Δtin2#3-tin2 were collected at 6 dpi. Excised leaf segments were ground in liquid nitrogen and served as a starting material for all subsequent analyses. (B) Measurement of total anthocyanin content. Anthocyanins were extracted and their amounts were measured at 515 nm using cyanidin 3-arabinoside chloride for calibration. Error bars indicate the standard deviation of three biological replicates. ***p<0.001 (vs SG200). (C) Specific accumulation of the three major anthocyanins: cyanidin 3-glucoside, cyanidin 3-(6″-malonylglucoside), and cyanidin 3-(3″,6″-dimalonylglucoside). Powdered samples were extracted and the polar phase was measured by UPLC-PDA-TOF-MS as described (Djamei et al., 2011). Data shown are representative of three biological replicates. ***p<0.001 (vs SG200). (D) Decreased accumulation of the three flavonoids in SG200Δtin2-infected tissue: orientin 2’’-O-rhamnoside, isoorientin 2’’-O-rhamnoside, and quercetin-glucoside-rhamnoside. Powdered samples were extracted and the polar phase was measured by UPLC-PDA-TOF-MS as described (Djamei et al., 2011). ***p<0.001 (vs SG200). (E) Decreased accumulation of DIBOA-glucoside in SG200Δtin2-infected tissue. Powdered samples were extracted and the polar phase was measured by UPLC-PDA-TOF-MS as described (Djamei et al., 2011). Data shown are representative of three biological replicates. *p<0.05, ***p<0.001 (vs SG200). (F) Decreased accumulation of ferulic acid in SG200Δtin2-infected tissue: powdered samples were extracted and the polar phase was measured by UPLC-PDA-TOF-MS as described (Djamei et al., 2011). Data shown are representative of three biological replicates. ***p<0.001 (vs SG200).
-
Figure 4—source data 1
Markers identified by metabolite fingerprinting (UPLC-ESI-TOF-MS analysis) in leaves of Z. mays 6 days post infection and verified by UHPLC-ESI-QTOF-MS/MS analysis or coelution.
- https://doi.org/10.7554/eLife.01355.010
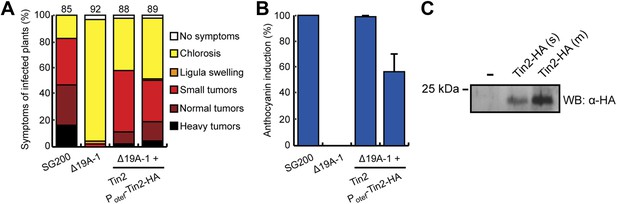
Biological activity of Tin2-HA and demonstration of its secretion.
SG200Δ19A-1-PotefTin2-HA expresses Tin2-HA protein under the constitutive otef promoter (Spellig et al., 1996). (A) Biological activity of Tin2-HA protein was confirmed by assaying virulence of SG200Δ19A-1-PotefTin2-HA and comparing this to SG200Δ19A-1-Tin2. (B) Complementation of anthocyanin induction by SG200Δ19A-1 strains by expressing Tin2-HA protein. The same infected plants as in (A) were scored for anthocyanin induction, which is given in % of all plants infected. (C) Detection of Tin2-HA protein in culture supernatants. Cells of SG200Δ19A-1 (−) and SG200Δ19A-1-PotefTin2-HA (s, single integration; m, multiple integration) were grown in CM liquid medium to an OD600 of 0.5. Proteins in supernatants were collected after TCA/DOC-precipitation and used in western blot analysis. The western blot was developed with anti-HA antibody (Sigma-Aldrich).
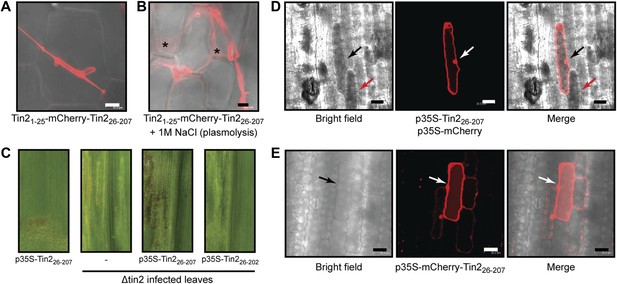
Tin2 protein secretion during biotrophic development and its function inside plant cells.
(A) Tin2 protein secretion during plant infection. Leaves are infected with SG200Δtin2-Tin21-25-mCherry-Tin226-207. mCherry fluorescence is observed at 3 dpi. Bar = 10 µm. (B) Same as (A) but treated with 1 M NaCl to induce plasmolysis. Asterisks mark enlarged apoplastic spaces. Bar = 10 µm. (C) Anthocyanin induction by transient expression of Tin226-207. Plasmid constructs indicated were delivered by biolistic gene transfer in maize leaves or leaves infected with SG200Δtin2. (D) Confocal microscopy of cells co-expressing Tin226-207 and mCherry. Dark staining indicates anthocyanin accumulation. Bar = 20 µm. (E) Confocal microscopy of mCherry-Tin226-207 transiently expressed in maize epidermal cells. The initially transformed cell is indicated by an arrow. Bar = 30 µm.
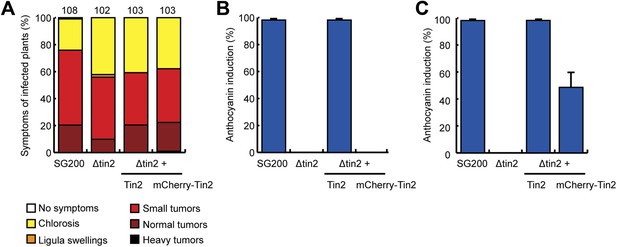
Biological activity of secreted mCherry-Tin226-207.
(A) Virulence of SG200Δtin2-Pcmu1Tin21-25-mCherry-Tin226-207 (labeled mCherry-Tin2) in comparison to SG200Δtin2. Disease symptoms were assessed at 12 dpi as described in legend to Figure 1—figure supplement 1. Tin21-25-mCherry-Tin226-207 was able to complement the virulence phenotype of SG200Δtin2. (B and C) The same infected plants as in (A) were scored at 6 dpi (B) and 12 dpi (C) for anthocyanin induction. Anthocyanin induction indicated in percent of total number of plants infected. Compared to SG200, SG200Δtin2-Pcmu1Tin21-25-mCherry-Tin226-207 was delayed and less efficient in anthocyanin induction.
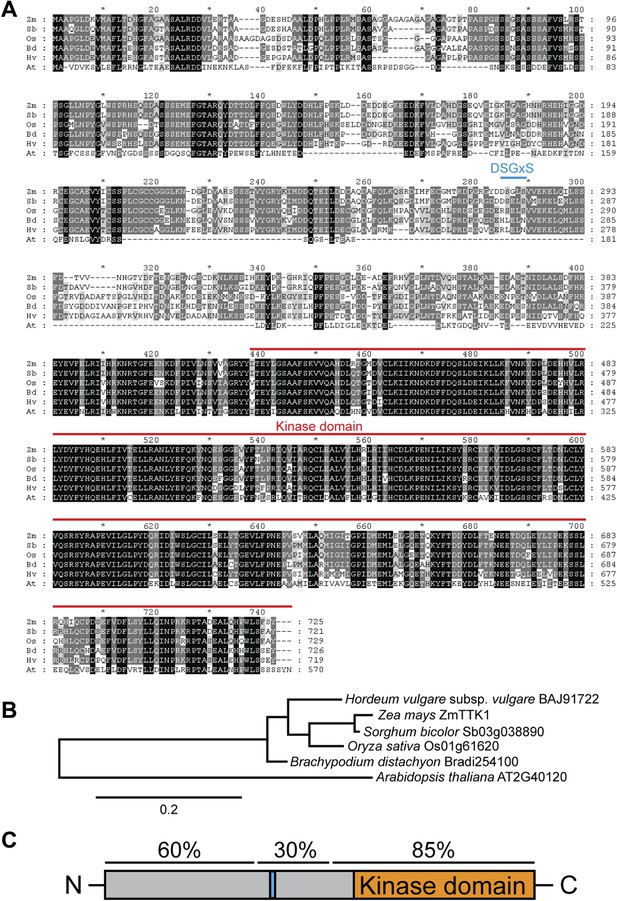
Comparison of ZmTTK1 orthologs from different grasses.
(A) Amino acid sequence alignment of ZmTTK1 orthologs. ZmTTK1 orthologs (Zm, Zea mays) could be identified in Sorghum bicolor (Sb), Oryza sativa (Os), Hordeum vulgare (Hv), Brachypodium distachyon (Bd) and a related protein found in Arabidopsis thaliana (At). Amino acids conserved in all 5 orthologs from monocot plants and the related protein in A. thaliana are highlighted in black, amino acids conserved in 4 or 5 of 6 these proteins are highlighted in dark gray and amino acids conserved in 3 of the 6 proteins are highlighted in light gray. The phosphodegron-like motif is indicated by a blue line. The kinase domain is indicated by a red line. (B) Phylogenetic tree of ZmTTK1 orthologs in monocot plants and the related protein from A. thaliana. Amino acid sequences of these proteins were analyzed by Phylogeny.fr to calculate the phylogenetic relationship. Bar indicates evolutionary distance. Accession numbers are indicated. (C) Schematic structure of ZmTTK1. The N-terminal region is labeled in gray and the C-terminal kinase domain labeled in orange. The phosphodegron-like motif is indicated in blue. Amino acid sequence identity of the N-terminal middle and C-terminal regions among orthologs in monocots is indicated above.
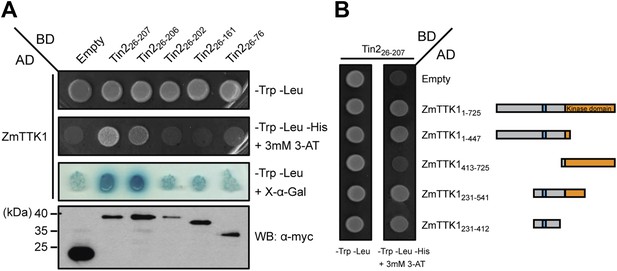
Mapping the interaction regions between Tin2 and ZmTTK1 by yeast two-hybrid analysis.
(A) A series of C-terminally truncated Tin2 proteins were expressed in fusion with GAL4BD (BD) protein and their interaction with full-length ZmTTK1 protein fused with GAL4AD (AD) were tested by yeast two-hybrid assay. Yeast transformants were grown on SD medium lacking indicated amino acids. Interaction was assessed from growth on SD -Trp -Leu -His + 3 mM 3-AT medium and from SD -Trp -Leu + X-α-Gal medium. Protein expression was verified by western blot with anti-myc antibody (Sigma-Aldrich) (bottom panel). (B) Mapping of the Tin2-interacting region of ZmTTK1. A series of truncated ZmTTK1 genes were fused with GAL4AD (AD) protein and their interaction with Tin226-207 protein fused with GAL4BD (BD) was assessed by yeast two-hybrid assay. Interaction was shown by growth of respective strains on SD -Trp -Leu -His + 3 mM 3-AT medium. Truncated ZmTTK1 versions are drawn schematically, the kinase domain is depicted in orange, and the N-terminal domain is colored in gray. The phosphodegron-like motif DSGxS is indicated in blue.
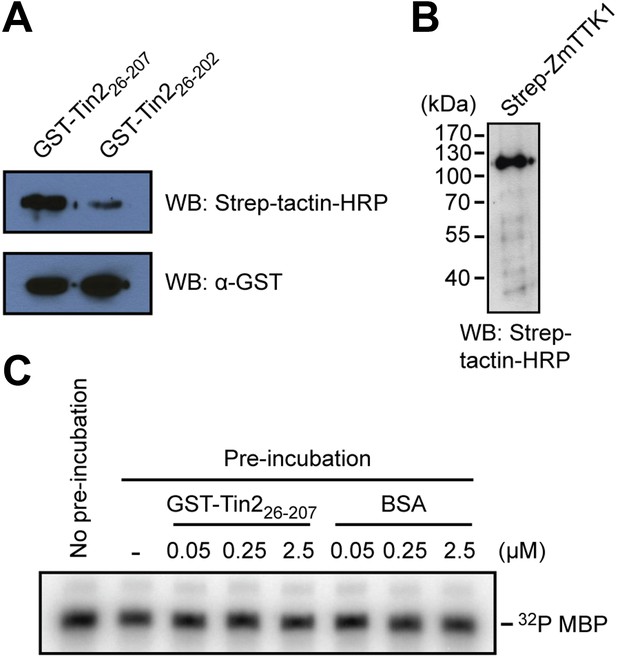
Physical interaction of Tin2 protein with full-length ZmTTK1 and in-vitro kinase activity of ZmTTK1.
(A) Physical interaction of Tin2 and ZmTTK1 demonstrated by in vitro GST-pull down assay using recombinant proteins. Recombinant GST-Tin226-207 or GST-Tin226-202 protein bound to glutathione sepharose beads (GE healthcare), respectively, was incubated with extract from induced BL21 (DE3)/pPRIBA102-ZmTTK1 at 4°C for 1 hr. GST fusion proteins were eluted with reduced glutathione. Strep-ZmTTK1 in eluate was detected by western blot (WB) using Strep-tactin-HRP and α-GST antibody for detection, respectively. Top panel detects precipitated Strep-ZmTTK1, bottom panel detects input GST-Tin2 fusion proteins. (B) Full-length recombinant Strep-ZmTTK1 expressed and purified from E. coli was detected by western blot analysis with Strep-tactin-HRP. (C) In vitro kinase activity of recombinant Strep-ZmTTK1 protein. Recombinant GST-Tin226-207 and Strep-ZmTTK1 proteins at indicated concentration were pre-incubated for 60 min at 4°C followed by addition of γ-32P ATP and myelin basic protein (MBP). Bovine serum albumin (BSA) was used as a control. Incubation continued at 28°C for 30 min. Proteins were separated by SDS-PAGE and phosphorylated protein was detected.
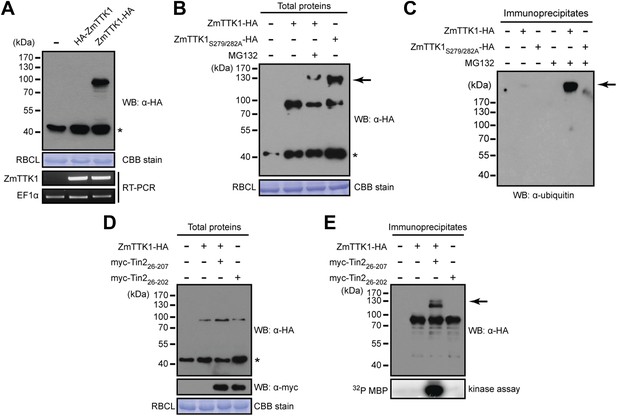
Tin2 protein stabilizes cytoplasmic maize protein kinase ZmTTK1.
(A) ZmTTK1-HA and HA-ZmTTK1 expression in N. benthamiana. HA-ZmTTK1 or ZmTTK1-HA protein was transiently expressed in N. benthamiana after infiltration with the respective A. tumefaciens strains GV3101 carrying pBIN19AN-HA-ZmTTK1 and pBIN19AN-ZmTTK1-HA. These plasmids express the indicated fusion proteins with a C-terminal IgG binding site. Expression was shown by western blot (WB) using indicated antibody. Asterisk labels a non-specific band. Rubisco large subunit (RBCL) stained with coomassie brilliant blue (CBB) served as a loading control. ZmTTK1-HA transcripts were analyzed by RT-PCR, EF1α served as a control. (B) Protein expression of ZmTTK1-HA with proteasome inhibitor MG132 (100 µM) and ZmTTK1S279/282A-HA. (C) Detection of poly-ubiquitinated ZmTTK1. After immunoprecipitation with human IgG-agarose, proteins were subjected to western blot to detect poly-ubiquitinated ZmTTK1 protein (arrow). The western blot was developed with monoclonal anti-ubiquitin antibody (Sigma-Aldrich). (D) Co-expression of ZmTTK1-HA with myc-Tin2. Total protein was analyzed by western blot using indicated antibodies. (E) Immunoprecipitated ZmTTK1-HA protein from (D) was analyzed by western blot. Kinase activity of immunoprecipitated samples shown on top was analyzed using MBP as a substrate (bottom).
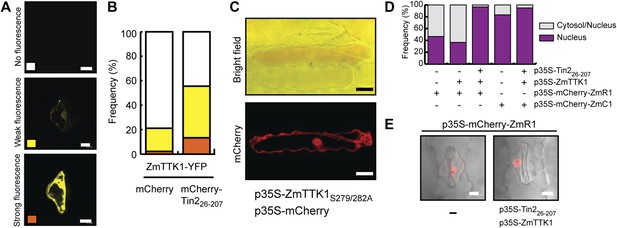
ZmTTK1 effects on localization of maize transcription factors and anthocyanin biosynthesis.
(A) Fluorescence intensity of ZmTTK1-YFP co-expressed with mCherry-Tin226-207 or with mCherry, respectively, in transiently transformed maize cells. Transformed maize cells were first identified by their mCherry fluorescence (not shown) and these cells were then screened for YFP fluorescence. The three examples depict the range of ZmTTK1-YFP expression patterns seen. (B) Quantification of the three YFP fluorescence patterns indicated in (A). Expressed proteins are listed below panels. In total, 90 transformed cells were scored for YFP fluorescence from three independent experiments. A statistically significant increase of weak and strong fluorescence patterns is observed when ZmTTK1-YFP is co-expressed with mCherry-Tin226-207 compared to co-expression of ZmTTK1-YFP with mCherry. (C) Anthocyanin induction after transient expression in maize. p35S-ZmTTK1S279/282A and p35S-mCherry were co-expressed in SG200Δtin2-infected leaves. Anthocyanin was visualized by bright field microscopy 3 days after particle bombardment. Bar = 20 µm. (D) mCherry-ZmR1 or mCherry-ZmC1 protein localization pattern. mCherry fluorescence in 60 cells from three different leaves was classified into cytoplasmic and nuclear localization or exclusive nuclear localization. (E) Confocal microscopy of mCherry-ZmR1. p35S-mCherry-ZmR1 was transiently introduced into maize either alone (−) or together with p35S-Tin226-207 and p35S-ZmTTK1. mCherry-ZmR1 was visualized. Bar = 10 µm.
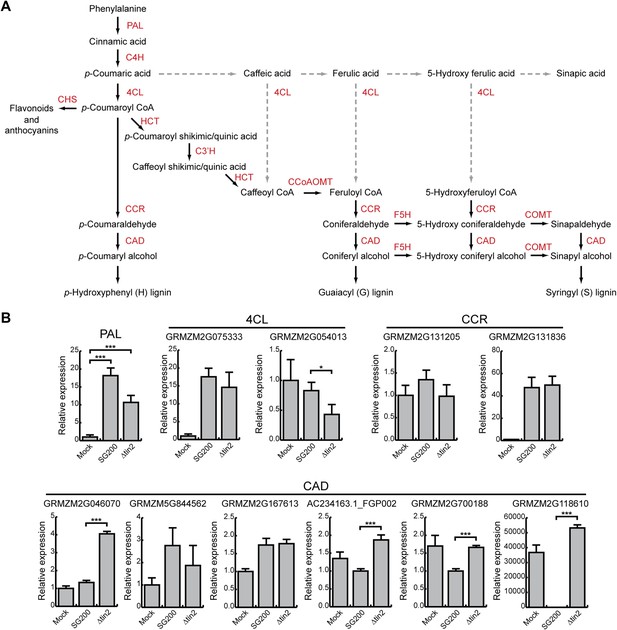
Phenylpropanoid pathway genes are differentially expressed after infection with SG200 and SG200Δtin2.
(A) Schematic model of the phenylpropanoid pathway in maize. The enzymes involved in each biosynthetic step are shown in red. PAL, phenylalanine ammonia lyase; C4H, cinnamate 4-hydroxylase; 4CL, 4-coumarate-CoA ligase; HCT, hydroxycinnamoyl transferase; C3′H, p-coumaroyl shikimate 3’-hydroxylase; CCoAOMT, caffeoyl CoA 3-O-methyltransferase; F5H, ferulate 5-hydroxylase; COMT, caffeic acid 3-O-methyltransferase; CCR, cinnamoyl-CoA reductase; CAD, cinnamyl alcohol dehydrogenase; CHS, chalcone synthase. Dashed lines indicate routes, which may occur in maize under certain conditions, but are not considered major routes based on experimental evidence (Humphreys and Chapple, 2002). (B) qPCR based quantification of the expression levels of genes from the phenylpropanoid pathway after infection with indicated U. maydis strains and collecting infected leaf material 6 dpi. *p < 0.05, ***p < 0.001.
-
Figure 12—source data 1
List of upregulated maize genes after SG200Δtin2 infection (vs SG200).
- https://doi.org/10.7554/eLife.01355.020
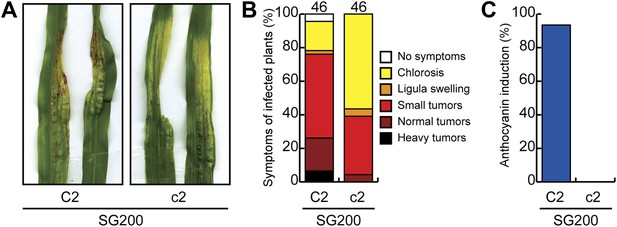
Pathogenicity of U. maydis in maize lacking chalcone synthase.
(A) Macroscopic SG200 disease symptoms on C2 (control) and c2 (chalcone synthase mutant) after 12 dpi are shown. (B) Disease symptoms were scored in two independent SG200 infections using the scoring scheme depicted on the right and the data were combined. (C) From the plants scored in (B) the percentage of plants showing anthocyanin induction is given.
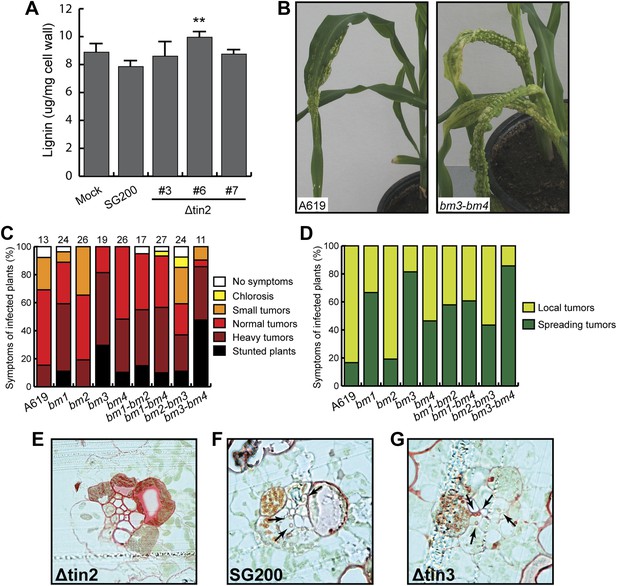
Virulence of U. maydis in hosts with altered lignin.
(A) Measurement of total lignin content in leaves infected with U. maydis. Infected leaf segments from mock inoculated leaves and leaves infected with three independent SG200Δtin2 strains as well as SG200 were collected at 6 dpi. Excised leaf segments were ground in liquid nitrogen and extracted with methanol. Isolated lignin was hydrolyzed under alkaline conditions and amount of monomers was measured at 280 nm using coniferyl alcohol for calibration. Error bars indicate the standard deviation of three biological replicates. **p<0.01 (vs SG200). (B) Macroscopic disease symptoms of U. maydis SG200 on maize A619 and the double brown midrib mutant bm3-bm4. Symptoms were scored at 12 dpi. (C) Pathogenicity of U. maydis SG200 on indicated bm mutants. Virulence was scored at 12 dpi. The total number of infected plants is indicated above each panel and data were combined from two independent experiments. (D) Symptoms of SG200 on bm mutants shown in (C) were classified into local tumors (tumors are restricted in one area on the leaf blade), and spreading tumors (tumors extend from infected leaf area all the way into the stem area). (E–G) Lignin deposition in cross-sections of plants infected with the indicated strains was visualized by Safranin O staining (red). Fungal hyphae inside vascular bundles are indicated by arrows.
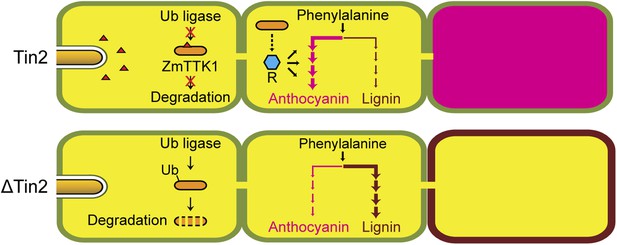
Hypothetical model for Tin2 function.
In the upper part the hypothetical model for infection of maize with wild-type U. maydis is depicted. Tin2 effector (red triangles) is secreted, taken up by maize cells and binding to the ZmTTK1 maize kinase (orange ellipsoid), which leads to its stabilization and renders it active. Active kinase is proposed to positively affect anthocyanin biosynthesis (dark pink) via the R transcription factor. Arrows and their thickness depict the activity of the anthocyanin and lignin pathway, respectively. The lower part depicts the hypothetical model for infection with an U. maydis tin2 mutant. In the absence of Tin2, ZmTTK1 is degraded via the ubiquitin-proteasome system. As a result, there is no anthocyanin biosynthesis, which could conceivably allow more precursor to enter the lignin branch, leading to fortified cell walls (brown lining) that limit fungal access.
Additional files
-
Supplementary file 1
Plasmids used in this study.
- https://doi.org/10.7554/eLife.01355.024
-
Supplementary file 2
Primers used in this study.
- https://doi.org/10.7554/eLife.01355.025
-
Supplementary file 3
Ustilago maydis strains used in this study.
- https://doi.org/10.7554/eLife.01355.026
-
Supplementary file 4
Saccharomyces cerevisiae strains used in this study.
- https://doi.org/10.7554/eLife.01355.027





