Transcription inhibition by the depsipeptide antibiotic salinamide A
Figures
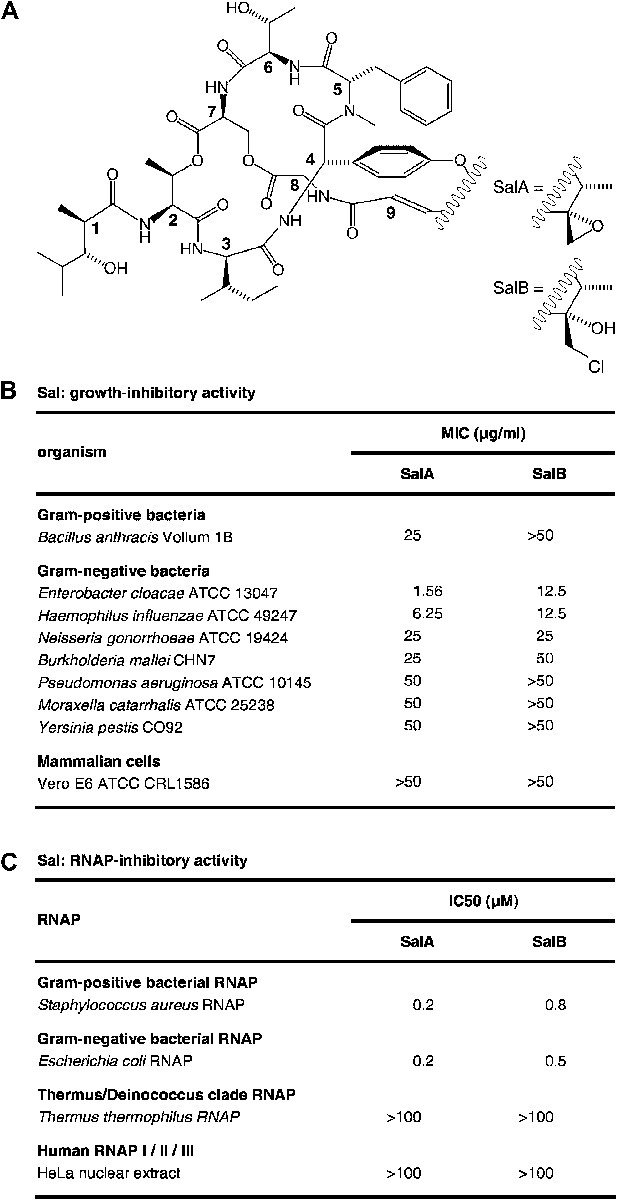
Sal.
(A) Structures of SalA and SalB (Moore et al., 1999). (B) Growth-inhibitory activity of SalA and SalB. (C) RNAP-inhibitory activity of SalA and SalB.
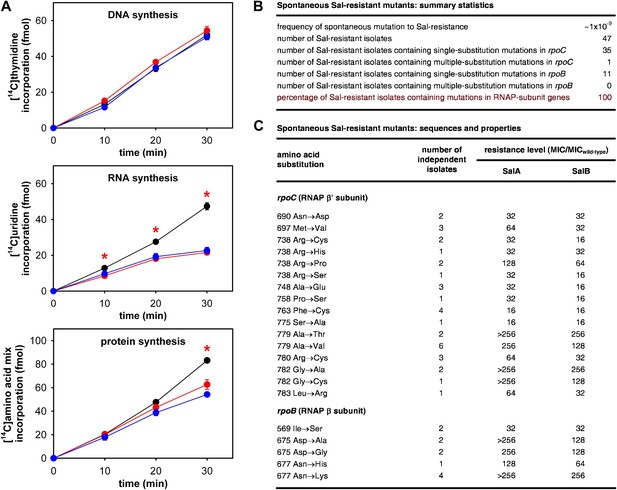
The RNAP-inhibitory activity of Sal accounts for the antibacterial activity of Sal.
(A) Sal inhibits RNAP in cells. Black, no inhibitor. Red, Sal (2 x MIC). Blue, Rif (2 x MIC). Asterisks, statistically significant differences between no-inhibitor data and Sal data (t test; p<0.01). (B and C) Sal-resistant mutations occur in RNAP subunit genes. MICwild-type,SalA = 0.049 µg/ml; MICwild−type,SalB = 0.20 µg/ml.
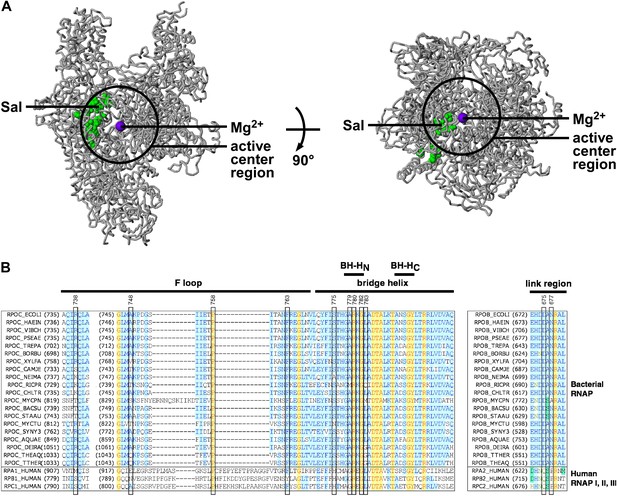
Target of transcription inhibition by Sal.
(A) The Sal target overlaps the RNAP active-center region. Structure of bacterial RNAP (gray ribbons; black circle for active-center region; violet sphere for active-center Mg2+; β' non-conserved region and σ omitted for clarity; PDB 1IW7), showing the sites of Sal-resistant substitutions (green surface; sequences from Figure 2C; ‘Sal target’). Two orthogonal views. (B) The Sal target overlaps the RNAP active-center module designated as the ‘bridge-helix cap’ (i.e., the module comprising the N-terminal half of the bridge helix, the F loop, and the link region; Weinzierl, 2010; Hein and Landick, 2010). Sequence alignments of the largest subunits of bacterial RNAP (top 20 sequences) and human RNAP I, RNAP II, and RNAP III (bottom three sequences), showing locations of Sal-resistant substitutions (black rectangles; sequences from Figure 2C; ‘Sal target’), and locations of the RNAP active-center bridge helix, bridge-helix N-terminal hinge (BH-HC), bridge-helix C-terminal hinge (BH-HC), F loop, and link region (black bars; boundaries from Weinzierl, 2010).
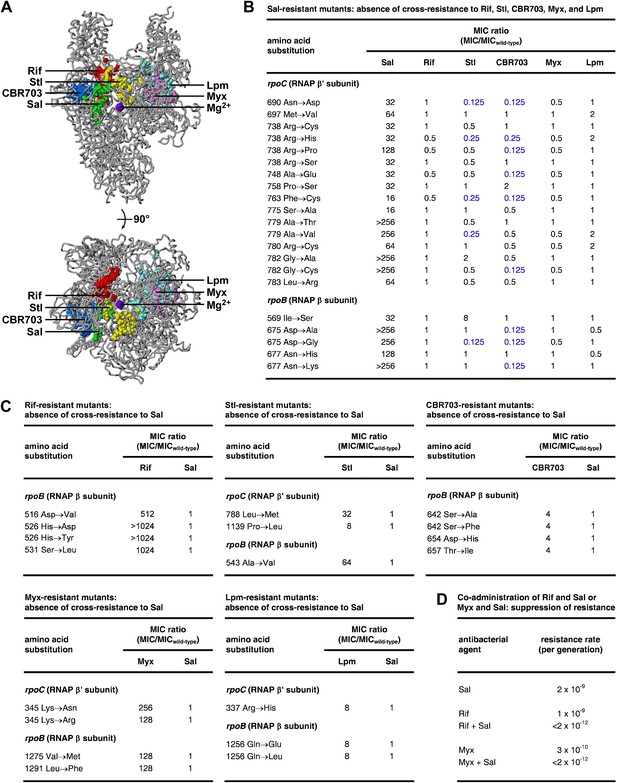
Relationship between the Sal target and the targets of other RNAP inhibitors.
(A) The Sal target does not overlap the targets of Rif, Stl, CBR703, Myx, and Lpm. Structure of bacterial RNAP (gray ribbons; violet sphere for active-center Mg2+; Mukhopadhyay et al., 2008), showing sites of substitutions that confer resistance to Sal (green; Figures 2, 3), Rif (red; Ovchinnikov et al., 1981, 1983; Lisitsyn et al., 1984; Jin and Gross, 1988; Severinov et al., 1993, 1994; Campbell et al., 2001; Garibyan et al., 2003), Stl (yellow; Lisitsyn et al., 1985; Heisler et al., 1993; Severinov et al., 1993, 1995; Tuske et al., 2005), CBR703 (blue; Artsimovitch et al., 2003; X Wang and RHE, unpublished), Myx (magenta; Mukhopadhyay et al., 2008; Srivastava et al., 2011), and Lpm (cyan; Ebright, 2005; Srivastava et al., 2011). Views as in Figure 3. (B) Sal-resistant mutants (Figure 2C) do not exhibit cross-resistance to Rif, Stl, CBR703, Myx, and Lpm. Blue, high-level hypersusceptibility (MIC ratio ≤0.25). MICwild-type,Sal = 0.049 μg/ml; MICwild-type,Rif = 0.20 μg/ml; MICwild-type,Stl = 3.13 μg/ml; MICwild-type,CBR703 = 6.25 μg/ml; MICwild-type,Myx = 0.20 μg/ml; MICwild-type,Lpm = 1.56 μg/ml. (C) Rif-resistant mutants (Jin and Gross, 1988; Garibyan et al., 2003; DD and RHE, unpublished), Stl-resistant mutants (Tuske et al., 2005), CBR703-resistant mutants (Artsimovitch et al., 2003; X Wang and RHE, unpublished), Myx-resistant mutants (Mukhopadhyay et al., 2008), and Lpm-resistant mutants (Ebright, 2005) do not exhibit cross-resistance to Sal. MICwild-type,Sal = 0.049 μg/ml; MICwild-type,Rif = 0.20 μg/ml; MICwild-type,Stl = 1.56 μg/ml; MICwild-type,CBR703 = 6.25 μg/ml; MICwild-type,Myx = 0.20 μg/ml; MICwild-type,Lpm = 1.56 μg/ml. (D) Co-administration of Sal (2 × MIC) with Rif (2 × MIC) or Myx (2 × MIC) suppresses the emergence of resistance.
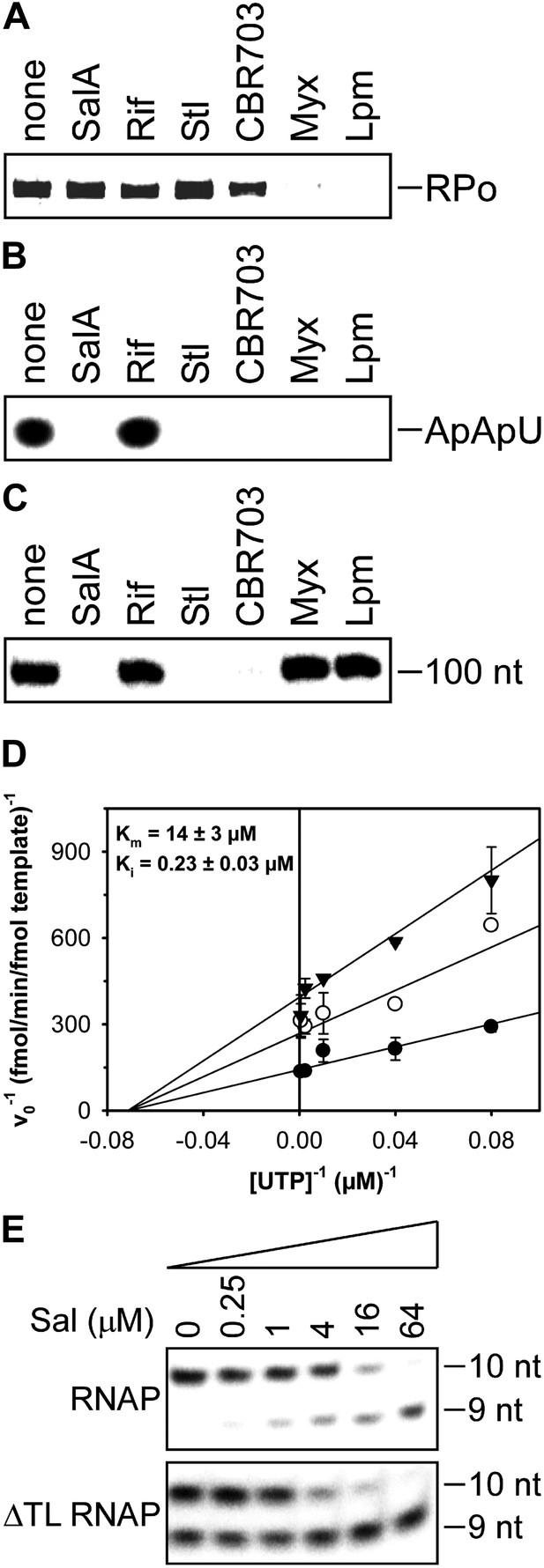
Mechanistic basis of transcription inhibition by Sal.
(A) Sal does not inhibit formation of a transcription initiation complex (RPo). (B) Sal inhibits nucleotide addition in transcription initiation. (C) Sal inhibits nucleotide addition in transcription elongation. (D) Sal inhibits nucleotide addition noncompetitively. (E) Transcription inhibition by Sal does not require the RNAP trigger loop.
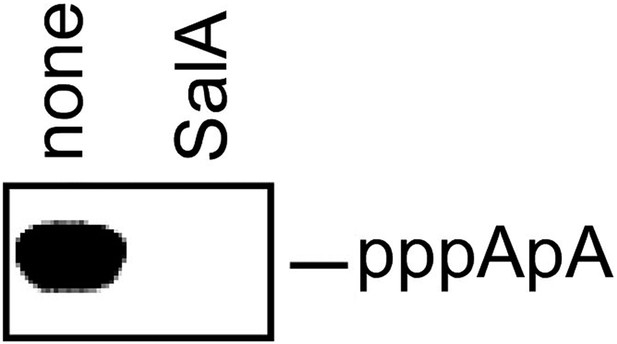
Sal inhibits nucleotide addition in de novo initiation.
https://doi.org/10.7554/eLife.02451.008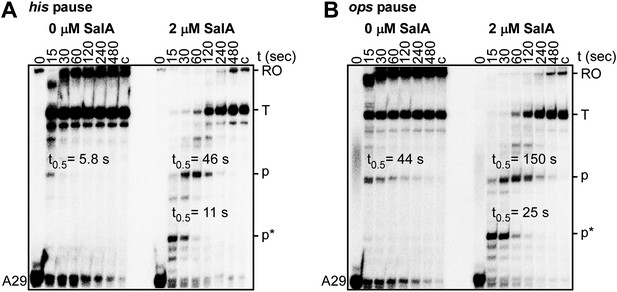
Sal enhances Type-I and Type-II transcriptional pausing.
(A) Effects of Sal on Type-I pausing at the his pause site. (B) Effects of Sal on Type-II pausing at the ops pause site. A29, 29 nt RNA product in halted elongation complex; c, ‘chase’ reaction; RO, run-off RNA product; T, terminated RNA product; p, ops or his pause-site RNA product; p*, additional pause-site RNA product; t0.5, pause half-life.
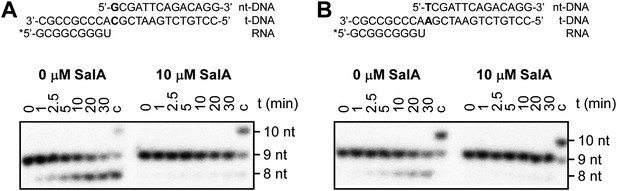
Sal inhibits pyrophosphorolysis.
(A) Pyrophosphorolysis assay using nucleic-acid scaffold containing G:C as first base pair of downstream duplex (relatively high pyrophosphorolysis rate in absence of inhibitor). (B) Pyrophosphorolysis assay using nucleic-acid scaffold containing T:A as first base pair of downstream duplex (relatively low pyrophosphorolysis rate in absence of inhibitor). Gel images show pyrophosphorolysis from 0 to 30 min. 9 nt, nucleic-acid scaffold; 8 nt, product of pyrophosphorolysis; 10 nt, product of ‘chase’ reaction with GTP (left) or UTP (right); nt-DNA, DNA nontemplate strand; t-DNA, DNA template strand.
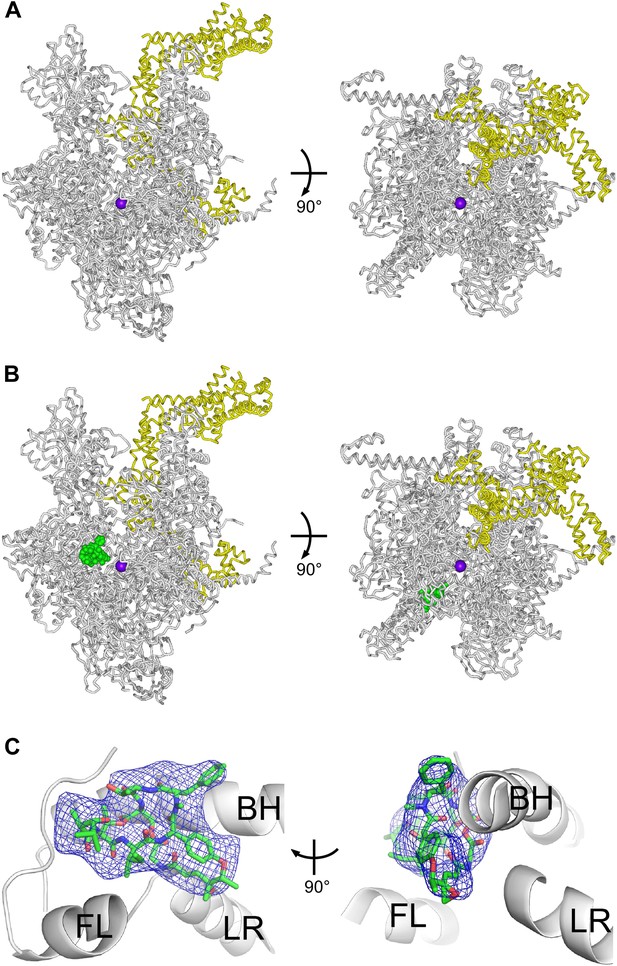
Structural basis of transcription inhibition by Sal: crystal structures of E. coli RNAP holoenzyme and E. coli RNAP holoenzyme in complex with Sal.
(A) Structure of E. coli RNAP holoenzyme (two orthogonal views). Gray ribbon, RNAP core. Yellow ribbon, σ70. Violet sphere, active-center Mg2+. (B) Structure of E. coli RNAP holoenzyme in complex with Sal (two orthogonal views). Green, Sal. Other colors as in A. (C) Electron density and atomic model for Sal (two orthogonal views). Blue mesh, NCS-averaged Fo-Fc omit map for Sal (contoured at 3.2σ). Green, red, and blue, Sal carbon, oxygen, and nitrogen atoms. Gray ribbons, RNAP. BH, FL, and LR, bridge helix, fork loop, and link region.
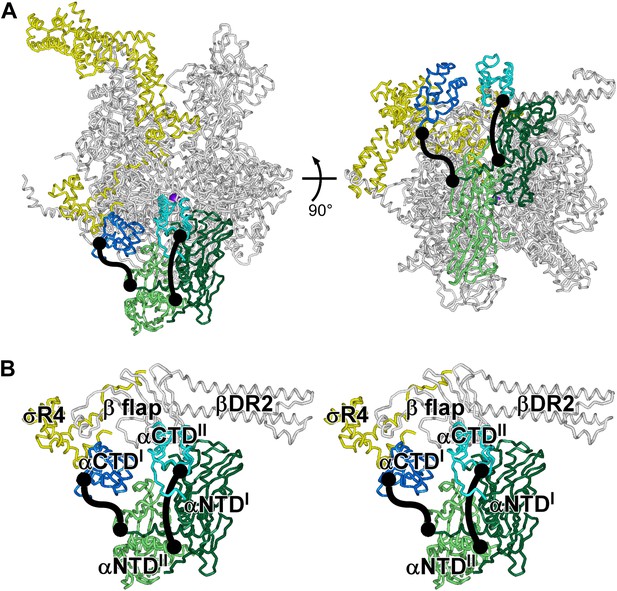
Structures of E. coli RNAP holoenzyme: αCTDI and αCTDII.
(A) Structure of E. coli RNAP holoenzyme (two orthogonal views). Gray, β', β and ω. Dark green and dark blue, αI subunit N-terminal and C-terminal domains (αNTDI and αCTDII). Light green and light blue, αII subunit N-terminal and C-terminal domains (αNTDII and αCTDII). Yellow, σ70. Violet sphere, active-center catalytic Mg2+. (B) Closeup view of αCTDI and αCTDII (stereoview). Gray, β flap and β dispensable region 2 (βDR2). Yellow, σ70 region 4 (σR4). Other colors as in A.
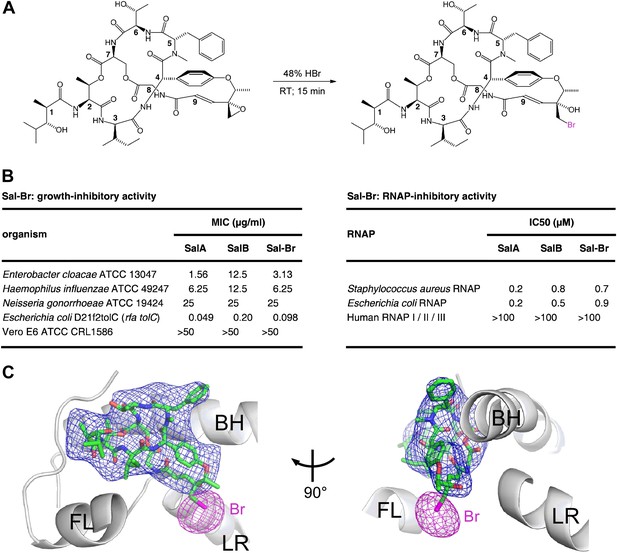
Structural basis of transcription inhibition by Sal: crystal structure of E. coli RNAP holoenzyme in complex with a bromine-containing Sal derivative.
(A) Synthesis of Sal–Br. (B) Growth-inhibitory activity and RNAP-inhibitory activity of Sal–Br. (C) Electron density, Br anomalous difference density, and atomic model for Sal–Br. Blue mesh, NCS-averaged Fo-Fc omit map for Sal (contoured at 3.2σ). Pink mesh, Br anomalous difference density for Sal–Br (contoured at 7.0σ). Other colors and labels as in Figure 6C.
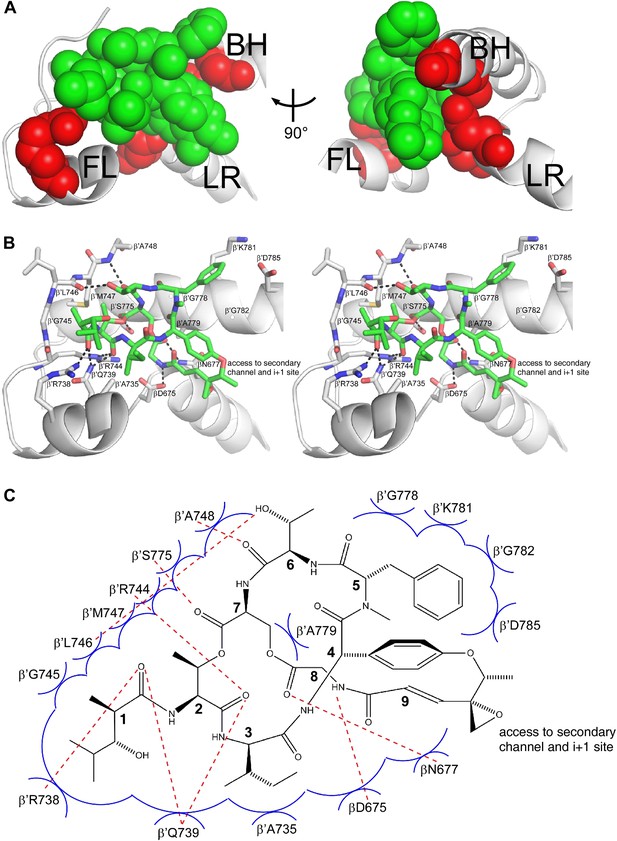
Structural basis of transcription inhibition by Sal: Sal makes direct interactions with the RNAP bridge-helix cap.
(A) Relationship between Sal (green) and sites of substitutions that confer high-level Sal-resistance (red). Views and labels as in Figures 6C and 7C. (B) Contacts between RNAP and Sal (stereoview). Gray, RNAP backbone (ribbon representation) and RNAP sidechain carbon atoms (stick representation). Green, Sal carbon atoms. Red, oxygen atoms. Blue, nitrogen atoms. Dashed lines, H-bonds. (C) Schematic summary of inferred contacts between RNAP and Sal. Red dashed lines, H-bonds. Blue arcs, van der Waals interactions.
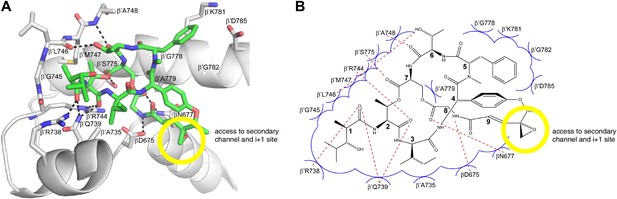
The structurally and chemically accessible epoxide moiety of SalA enables semi-synthesis of novel Sal analogs.
Yellow circle, Sal residue-9 epoxide moiety. Other colors as in Figure 8B,C.
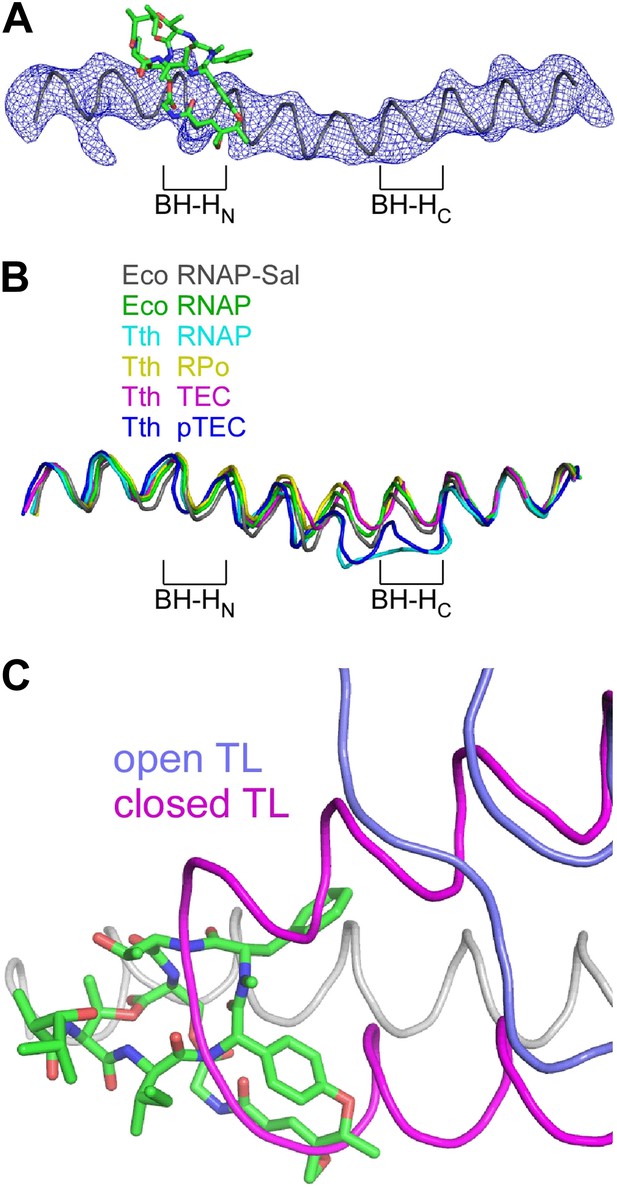
Structural basis of transcription inhibition by Sal: Sal interacts with an ‘open’ (unbent) state of the bridge-helix N-terminal hinge and an ‘open’ (unfolded) state of the trigger loop.
(A) Electron density and model for bridge helix in crystal structure of RNAP-Sal. Blue mesh, Fo-Fc omit map for bridge helix (contoured at 2.5σ). Black ribbon, bridge-helix backbone. Green, red, and blue, Sal carbon, oxygen, and nitrogen atoms. BH-HN, bridge-helix N-terminal hinge. BH-HC, bridge-helix C-terminal hinge. (B) Superimposition of bridge helices of E. coli RNAP-Sal (black), E. coli RNAP (green; unbent BH-HN and BH-HC), T. thermophilus RNAP (cyan; PDB 1IW7), T. thermophilus RPo (yellow; PDB 4G7H), T. thermophilus transcription elongation complex (pink; PDB 2O5J), and paused T. thermophilus transcription elongation complex (violet; PDB 4GZY). (C) Predicted absence of steric clash between Sal (colors as in A) and trigger loop in open conformational state (blue; PDB 1ZYR) and predicted presence of steric clash between Sal and trigger loop in closed conformational state (pink; PDB 2O5J).
Additional files
-
Supplementary file 1
Plasmid-borne induced Sal-resistant mutants: sequences and properties.
- https://doi.org/10.7554/eLife.02451.017
-
Supplementary file 2
Crystal structures of E. coli RNAP holoenzyme and E. coli RNAP holoenzyme in complex with Sal: crystallization and refinement statistics.
- https://doi.org/10.7554/eLife.02451.018
-
Supplementary file 3
Crystal structure of E. coli RNAP holoenzyme in complex with a bromine-containing Sal derivative: crystallization and refinement statistics.
- https://doi.org/10.7554/eLife.02451.019





