Specific polar subpopulations of astral microtubules control spindle orientation and symmetric neural stem cell division
Figures
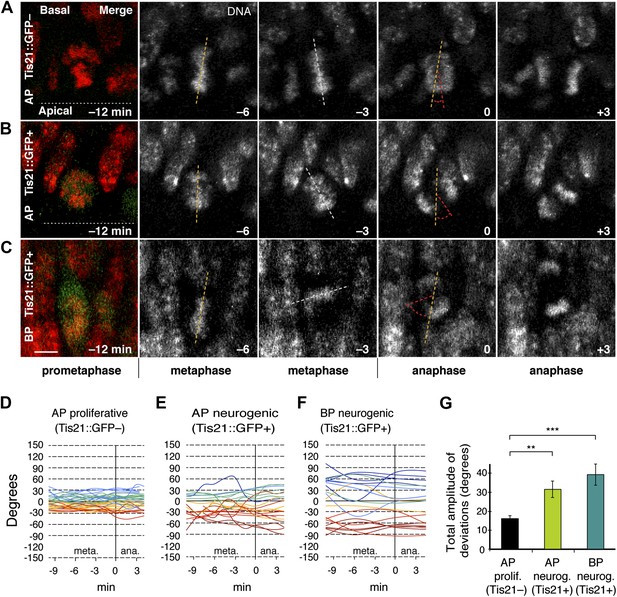
Dynamic spindle orientation variability increases when progenitors become neurogenic.
Live tissue imaging of spindle orientation as reported by chromosome plate orientation in organotypic slice culture of coronal sections from E14.5 Tis21::GFP mouse dorsolateral telencephalon. Observations focused on metaphase, when all chromosomes congressed to the equatorial plane and the dynamics observed were likely more related to spindle orientation, rather than the prometaphase establishment of a functional spindle and chromosome plate. 0 min is anaphase onset. (A–C) Apical progenitor (AP; A, B) and basal progenitor (BP; C) undergoing either proliferative (A, Tis21::GFP–) or neurogenic (B, C; Tis21::GFP+) division. Merge: chromosomes in red, EGFP in green. Chromosome plate orientation was determined by measuring angular deviations from the apico-basal axis (0°, corresponding to vertical), which runs perpendicular to the apical ventricular surface (90°, horizontal white dotted lines in A, B). Time-lapse is 3 min. Vertical or oblique dashed lines indicate chromosome plate orientations in metaphase or anaphase onset; maximal deviation angles for each plate were set by the orientation at an early time-point (yellow) and a later time-point (red), which are quantified in G. White dashed lines indicate intermediate orientations. Scale bar = 5 μm. (D–F) Quantification of chromosome plate orientations from metaphase (meta.) to anaphase (ana.). To facilitate tracing, individual tracks are colour-coded according to the range in which most of the track remained (blue, beyond 30°, cyan 30°–15°, green 15°–0°, yellow 0° to −15°, red −15° to −30°, dark red, beyond −30°). (G) Mean ± standard error of the mean (SEM) of the maximal amplitude of deviations for proliferative (prolif.) and neurogenic (neurog.) divisions. **p<0.001, ***p<0.0001, Kruskal–Wallis ANOVA (K–W) with Dunn's multiple comparison (DMC) test; n = 20 progenitors per category, from three independent litters and experiments. See also Videos 1–3.
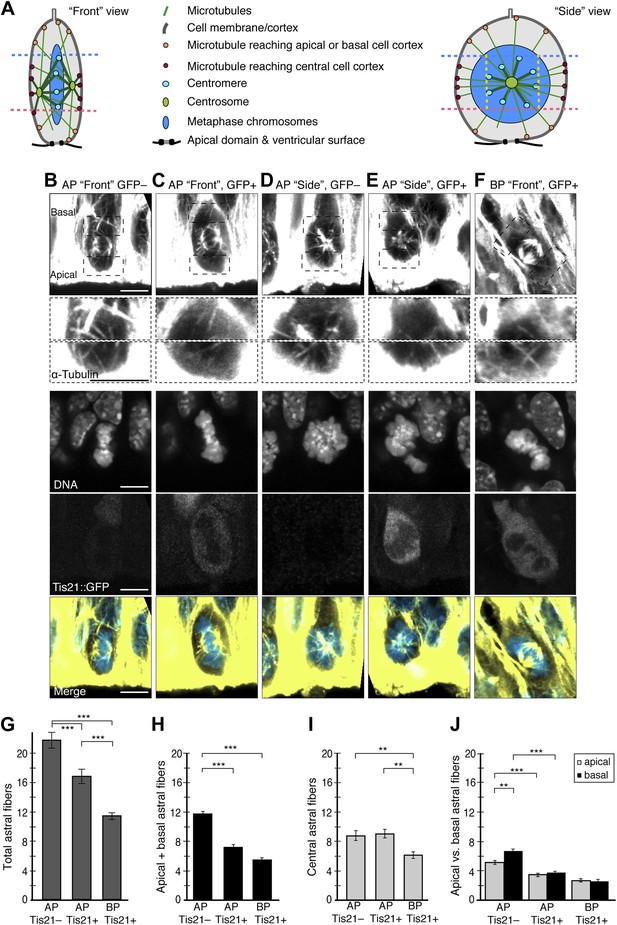
Fewer astral microtubules when progenitors become neurogenic.
(A) Cartoons of the mitotic spindle in APs as appearing upon ‘Front’ and ‘Side’ viewing of the same chromosome plate, rotating around its apico-basal axis. Apical- and basal-reaching astrals (apical/basal astrals) were defined as those extending beyond the main chromosomal area, delimited by the apical-most (pink dashed line) and basal-most (blue dashed line) peri/centromeric heterochromatin foci (brightest points in DNA, see B–F), and reaching either the apical or basal region of the cell cortex, which are delimited by these same pink and blue dashed lines, respectively. In the ‘Side’ view, yellow dashed lines delimit the peri/centromeric heterochromatin region laterally, beyond which the astrals defined as central-reaching (central astrals) extend. (B–F) α-tubulin immunofluorescence (maximum intensity projections of two 0.75 μm optical sections) of E14.5 Tis21::GFP mouse dorsolateral telencephalon showing mitotic microtubules in proliferating APs (Tis21::GFP–) vs neurogenic APs (Tis21::GFP+) vs neurogenic BPs (Tis21::GFP+). Dashed boxes show regions including the basal and apical cell cortex that are shown in the second and third row, respectively, at higher magnification and brightness. DNA staining (DAPI) and Tis21::GFP fluorescence are single 0.75 μm optical sections. Merge: DNA in blue, microtubules in yellow. Scale bars = 5 μm. (G–J) Mean numbers per cell of astrals reaching the cell periphery: all astrals (G), apical/basal astrals (H), central astrals (I), and apical or basal astrals (J). (G, H, J) In BPs, astrals are considered as apically, basally, and centrally oriented (see ‘Materials and methods’). n = 40 progenitors per category and I n = 20 progenitors per category, all from 8 independent litters and experiments. *p<0.05, **p<0.001, ***p<0.0001; K–W with DMC post test. Error bars are SEM.
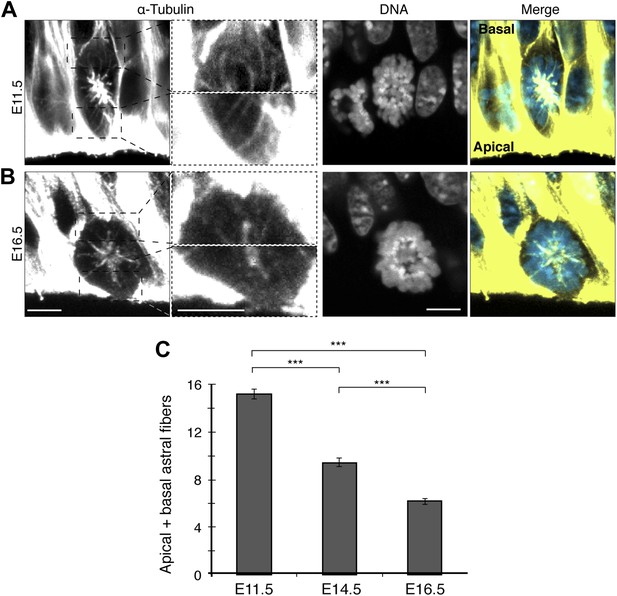
Fewer apical/basal astral microtubules in progenitors as neurogenesis progresses.
A, B) α-Tubulin immunofluorescence (maximum intensity projections of two 0.75 μm optical sections) of E11.5 and E16.5 mouse dorsolateral telencephalon showing mitotic microtubules in APs. Dashed boxes in the first column show regions including the basal and apical cell cortex that are shown at higher magnification and brightness in the second column. DNA stainings (DAPI) are single 0.75 μm optical sections. Merge: DNA in blue, microtubules in yellow. Scale bars = 5 μm. (C) Mean number per cell of apical/basal astrals in APs at E11.5, E14.5 (Figure 2H, mean of both AP categories), and E16.5. ***p<0.0001, K–W with DMC post test; for E11.5 and E16.5, n = 20 APs, from 4 independent litters and experiments; Error bars are SEM.
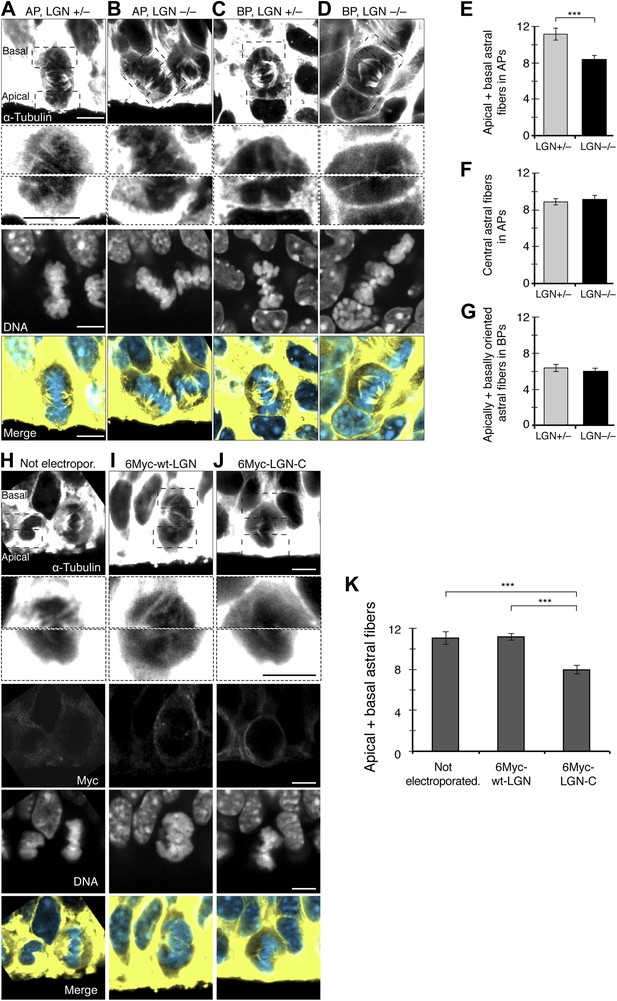
Fewer astral microtubules in LGN KO progenitors and upon dominant-negative LGN-C overexpression.
(A–D) α-Tubulin immunofluorescence (maximum intensity projections of two 0.75 μm optical sections) of E14.5 heterozygous (LGN +/−) vs homozygous (LGN −/−) E14.5 LGN KO mouse dorsolateral telencephalon showing mitotic microtubules in APs and BPs. Dashed boxes show regions including the basal and apical cell cortex that are shown in the second and third row, respectively, at higher magnification and brightness. DNA staining (DAPI) and Tis21::GFP fluorescence are single 0.75 μm optical sections. Merge: DNA in blue, microtubules in yellow. Scale bars = 5 μm. (E) Mean number per AP of astrals reaching the apical and basal cell cortex (apical/basal astrals). (F) Mean number per AP of astrals reaching the central cell periphery (central astrals). (G) Mean number per BP of astrals that are apically and basally oriented. ***p=0.0007, one-tail t-test, n (20 progenitors per category from 3 independent litters and experiments). Error bars are SEM. See also Figure 5. (H–J) α-Tubulin immunofluorescence (maximum intensity projections of two 0.75 μm optical sections) of in utero electroporated E13.5 wt mouse dorsolateral telencephalon analysed 24 hr later (E14.5). APs in the electroporated regions were either not electroporated (H), or electroporated with LGN-wt (6Myc-wt-LGN, I) or with dominant-negative LGN (6Myc-LGN-C, J). Dashed boxes in the first column show regions including the basal and apical cell cortex that are shown at higher magnification and brightness in the second column. DNA staining (DAPI) and Myc immunofluorescence are single 0.75 μm optical sections. Merge: DNA in blue, microtubules in yellow. Scale bars = 5 μm. (K) Mean number per cell of astral microtubules reaching the apical or basal cell cortex (apical + basal astrals) of APs in the electroporated region that were either not electroporated, or transfected with 6Myc-LGN-wt or with 6Myc-LGN-C. ***p<0.0001, one-way ANOVA with TMC test; n = 20 progenitors per category, from 3 independent litters and experiments. Error bars are SEM. Scale bars = 5 μm.
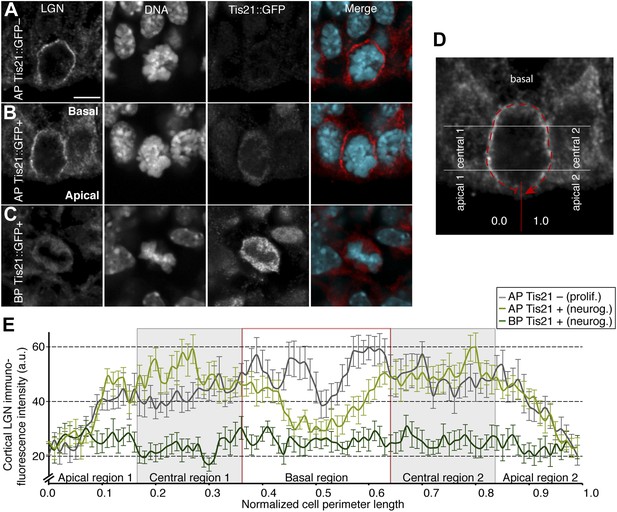
Less cell cortical LGN in neurogenic progenitors.
(A–C) Double immunofluorescence for LGN and GFP, with DNA staining (DAPI), of E14.5 Tis21::GFP mouse dorsolateral telencephalon showing representative examples of a proliferating AP (Tis21::GFP–) vs neurogenic AP (Tis21-GFP+) vs neurogenic BP (Tis21-GFP+). Merge: DNA in cyan, LGN in red. (D) Same mitotic AP as in (I), illustrating how LGN cortical immunofluorescence intensity was measured, starting at 0.0 in the middle of the apical region and continuing clockwise along the entire cell cortex (dashed red line) until 1.0. Apical, central, and basal regions are indicated (regions 1, left side; regions 2, right side). (E) Mean ± SEM of LGN cortical immunofluorescence intensity along 100 equidistant points of the normalized cell perimeter length; the light grey boxes highlight the central regions; the red box highlights the basal region; n = 15 progenitors per category, from 4 independent litters and experiments. Scale bars = 5 μm.
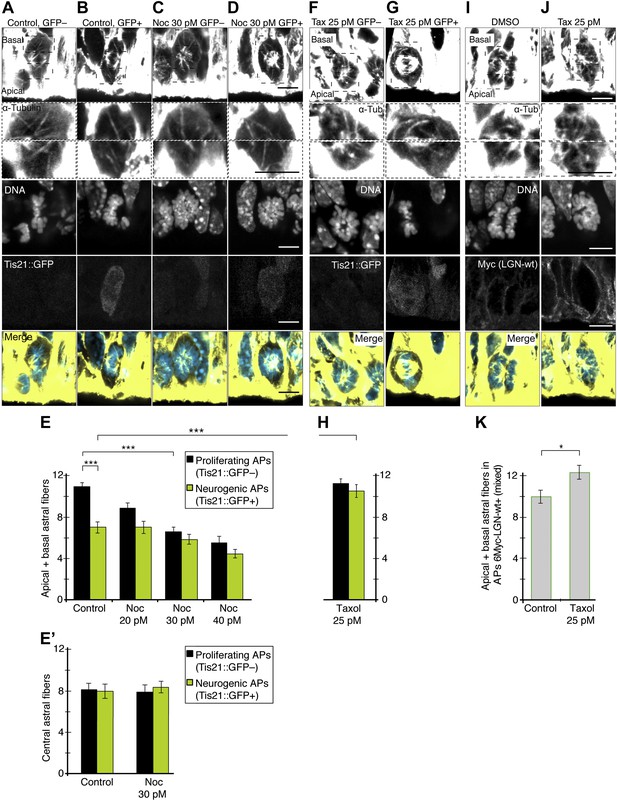
Changing the number of apical/basal astral microtubules in APs.
A–E) Controlled decrease in the apical/basal astrals of proliferating APs under minimal nocodazole. E14.5 forebrains from Tis21::GFP mice were incubated for 2.5 hr in BRO culture, either with solvent (DMSO) only (Control) or with 30 pM nocodazole (Noc). (A–D) α-Tubulin immunofluorescence (maximum intensity projections of two 0.75 μm optical sections) of dorsolateral telencephalon showing mitotic microtubules in proliferating APs (Tis21::GFP–) vs neurogenic APs (Tis21::GFP+), either in controls or with 30 pM nocodazole. Dashed boxes show regions including the basal and apical cell cortex that are shown in the second and third row, respectively, at higher magnification and brightness. DNA staining (DAPI) and Tis21::GFP fluorescence are single 0.75 μm optical sections. Merge: DNA in blue, microtubules in yellow. Scale bars = 5 μm. (E) Mean number per cell of apical/basal astrals in control or upon incubation with increasing concentrations of nocodazole in the picomolar range. Note that, with 30 pM nocodazole, the mean number of apical/basal astrals of proliferating APs (Tis21::GFP–) was both significantly lower than in control proliferating APs and similar to that of control neurogenic APs (Tis21::GFP+); ***p<0.0001, K–W with DMC post test; n = 20 progenitors per category, from 3 independent litters and experiments. (E') Mean number per cell of central astrals in controls or with 30 pM nocodazole. n = 10 progenitors per category, from 3 independent litters and experiments. Error bars are SEM. See also Figure 6—figure supplement 1. (F–K) Controlled increase in apical/basal astrals of neurogenic APs under minimal taxol and also in APs overexpressing LGN-wt combined with minimal taxol. (F, G) E14.5 forebrains from Tis21::GFP mice were incubated for 2.5 hr in BRO culture with 25 pM taxol. α-Tubulin immunofluorescence (maximum intensity projections of two 0.75 μm optical sections) of dorsolateral telencephalon showing mitotic microtubules in a proliferating AP (Tis21::GFP–) vs a neurogenic AP (Tis21::GFP+). Dashed boxes in the first row show regions including the basal and apical cell cortex that are shown at higher magnification and brightness in the second and third rows. Tis21::GFP fluorescence (fifth row) and DNA staining (DAPI, fourth row) are single 0.75 μm optical sections. Merge: DNA in blue, microtubules in yellow. Scale bars = 5 μm. (H) Mean number per cell of apical/basal astrals upon 25 pM taxol incubation. The mean number of apical/basal astrals of neurogenic APs (Tis21::GFP+) was both significantly higher than in control neurogenic APs (E) and similar to that of control (E) and taxol-treated (H) proliferating APs (Tis21::GFP–). n = 12 progenitors per category, from three independent litters and experiments. (I, J) E13.5 wt forebrains were subjected to in utero electroporation of 6Myc-LGN-wt and 18 hr later (E14.25) incubated for 2.5 hr in BRO culture with either DMSO only (control) or 25 pM taxol. Immunofluorescences similar to F, G, but the fifth row shows immunofluorescence for Myc (6Myc-LGN-wt). (K) Mean number of apical/basal astrals of APs electroporated with 6Myc-LGN-wt. This was higher upon taxol incubation than in controls. *p<0.05, ***p<0.0001, K–W with DMC post test; n = 12 progenitors per category, from n ≥ 2 independent litters and experiments. Error bars are SEM.
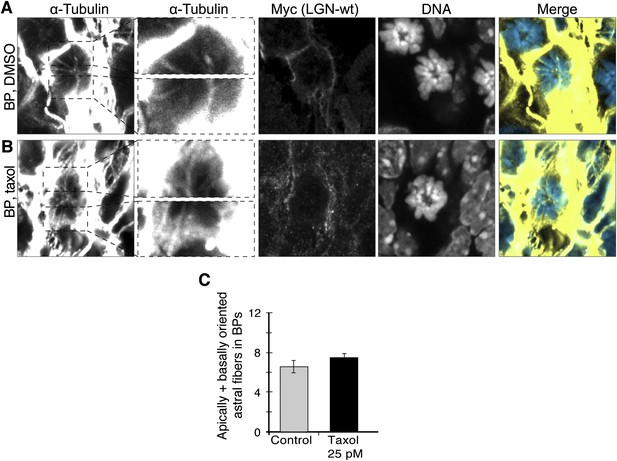
No increase in apical/basal astral microtubules of BPs upon wt LGN overexpression combined with minimal taxol incubation.
E13.5 wt forebrains were subjected to in utero electroporation of 6Myc-LGN-wt and 18 hr later (E14.25) incubated for 2.5 hr in BRO culture with either solvent (DMSO) only (Control) or with 25 pM taxol. (A, B) α-Tubulin and Myc (6Myc-LGN-wt) immunofluorescence (maximum intensity projections of two 0.75 μm optical sections) of dorsolateral telencephalon showing mitotic microtubules in BPs. Dashed boxes in the first column show regions including the basal and apical cell cortex that are shown at higher magnification and brightness in the second column. DNA stainings (DAPI, fourth column) are single 0.75 μm optical sections. Merge: DNA in blue, microtubules in yellow. Scale bars = 5 μm. (C) Mean ± SEM of apically and basally oriented astral microtubules per BP upon incubation with DMSO only (control) or 25 pM taxol. The mean number of apically and basally oriented astral microtubules was not significantly different between control and taxol-incubated BPs; n ≥ 20 progenitors per category, from two independent litters and experiments.
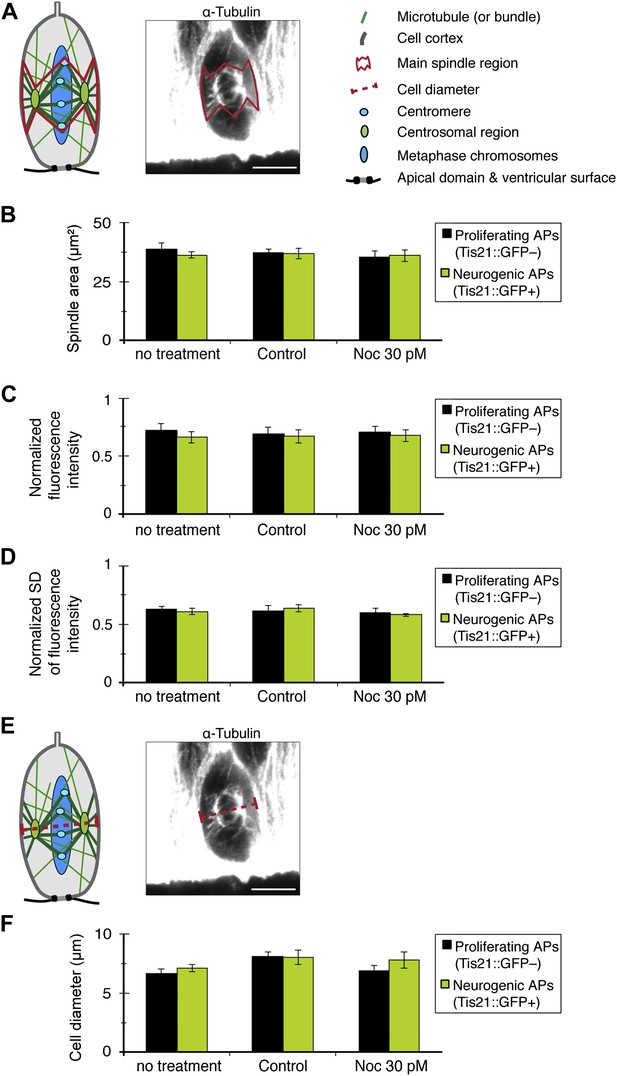
Key spindle and cell features in APs are not altered by 30 pM nocodazole.
All measurements were performed on the same cell images from which the astral microtubule data were obtained (see Figures 2 & 4). (A) Cartoon and example image (α-tubulin immunofluorescence) of the mitotic spindle in an AP, indicating the main spindle region (solid red line) where α-tubulin immunofluorescence measurements were performed to control for variations in spindle features (B–D), between proliferating and neurogenic APs, either without (no treatment) or with BRO culture of Tis21::GFP E14.5 forebrains, with either solvent (DMSO) only (Control) or with 30 pM nocodazole (Noc). The main spindle region was defined on the basis of the cell body shape and the centrosomal region (both revealed by α-tubulin immunofluorescence) and the apical- and basal-most (peri-)centromeric foci (DAPI staining) (see Figure 2A). The α-tubulin immunofluorescence used for illustration on the right side in A and E is the same as in Figure 2C. The key in A applies also for the cartoon in E. To obtain values from the central part of the spindle, the mean of the measurements from the 2 central-most 0.75 μm confocal sections was obtained. (B) Quantification of the area of the main spindle region. (C) Quantification of the amount of spindle microtubules, as obtained from the mean fluorescence intensity in the main spindle region normalized to the mean fluorescence intensity in the entire image. (D) Spatial distribution of α-tubulin in the main spindle region, as revealed by the mean standard deviation (SD) between the per-pixel fluorescence intensity values in the main spindle region, normalized to the mean fluorescence intensity of the main spindle region. (E) Cartoon and example image of the mitotic spindle showing the AP diameter measurement at the spindle plane (red dashed line). (F) AP cell body diameter at the spindle plane, indicating also spindle length. (B–D, F) Significant differences were not found with K–W with DMC post test. Error bars are SEM. Scale bars = 5 μm. See also Figure 7—source data 1 and Figure 7—figure supplement 1.
-
Figure 7—source data 1
Table with the values for the graphs in Figure 7.
- https://doi.org/10.7554/eLife.02875.014
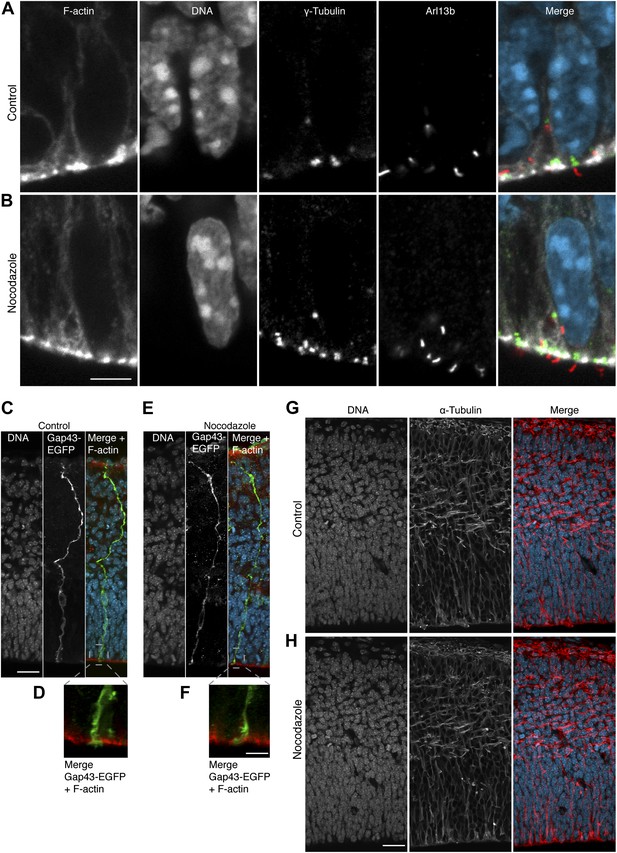
Key subcellular, cellular, and tissue features in APs are not significantly altered by 30 pM nocodazole.
E14.5 forebrains from wt mice were incubated for 2.5 hr in BRO culture either with solvent (DMSO) only (control) or with 30 pM nocodazole, and immunofluorescences of dorsolateral telencephalon sections were performed. (A, B) Maximum intensity projections of two 0.9 μm optical sections showing F-actin (phalloidin, white), DNA (DAPI, cyan), γ-tubulin (green) as centriolar marker and Arl13b (red) as ciliary marker, showing representative examples of primary cilia protruding from the apical domain (negative F-actin staining) with normal architecture in controls or with nocodazole. Scale bar = 5 μm. (C–F) Maximum intensity projections of four 0.9 μm optical sections of GAP43-GFP (green) showing representative examples of elongated APs, from basal (top) to apical (bottom), together with DNA (DAPI, cyan) and F-actin (phalloidin, red), with normal architecture in controls or with nocodazole. Scale bar = 20 μm. (D, F) Zoom-ins of the apical region (dashed grey boxes in the merges in C and E, but without DNA), with the apical end-foot of the APs embedded in the apical ventricular surface, in DMSO and nocodazole, respectively. Scale bar = 5 μm. (G, H) Maximum intensity projections of two 0.9 μm optical sections of α-tubulin (red) and DNA (cyan), showing normal cytoarchitecture of the cortical wall as revealed by the pattern of microtubules in representative coronal sections of the entire developing cortical wall of controls or with nocodazole. Scale bar = 20 μm; n = 3 brains from 2 independent litters and experiments per category.
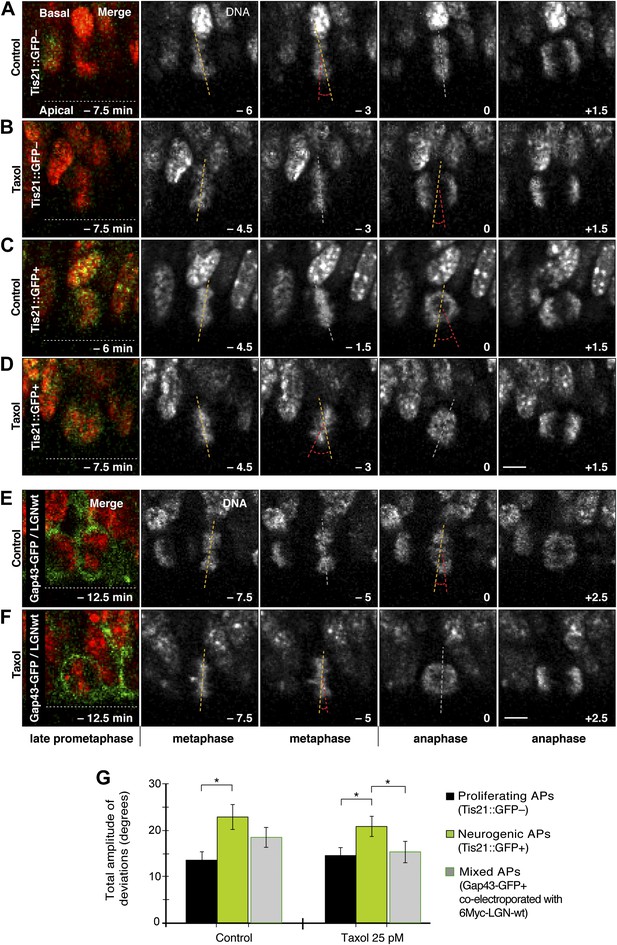
Dynamic spindle orientation variability decreases with minimal taxol incubation combined with LGN-wt overexpression, but not with taxol alone.
A–D) E14.5 forebrains from Tis21::GFP mice were incubated on the microscope stage with either solvent (DMSO) only (Control) or with 25 pM taxol. Live tissue imaging of spindle orientation, as reported by chromosome plate orientation, in organotypic slice culture of coronal sections from Tis21::GFP mouse dorsolateral telencephalon. Time-lapse is 1.5 min. 0 min corresponds to anaphase onset. Apical progenitors underwent either proliferative (A, B, Tis21::GFP–) or neurogenic (C, D; Tis21::GFP+) divisions. Merge: chromosomes in red, EGFP in green. Chromosome plate orientation was determined by measuring angular deviations from the apico-basal axis (0°, corresponding to vertical), which runs perpendicular to the apical ventricular surface (90°, horizontal white dotted lines in A–F). Vertical or oblique dashed lines indicate chromosome plate orientations in metaphase or anaphase onset; maximal deviation angles for each plate were set by the orientation at an early time-point (yellow) and a later time-point (red), which are quantified in G. White dashed lines indicate intermediate orientations. Scale bar = 5 μm. (E, F) Similar to A,B and C,D, but with E13.5 wt forebrains subjected to in utero electroporation of 6Myc-LGN-wt together with GAP43-EGFP and 18 hr later (E14.25) incubated for 2.5 hr in organotypic slice culture with either DMSO only (control) or 25 pM taxol. Time-lapse is 2.5 min. (G) Mean ± SEM of the maximal amplitude of deviations for proliferating (Tis21::GFP–) vs neurogenic (Tis21::GFP+) APs or of APs electroporated with LGN-wt together with GAP43-EGFP, in control or 25 pM taxol incubations. *p<0.05, K–W with DMC post test, n = 20 progenitors per category, from two independent litters and experiments.
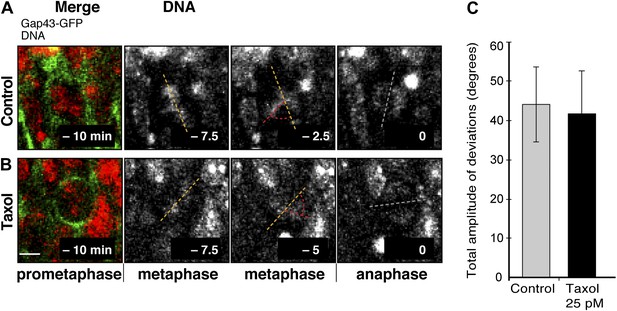
No increase in dynamic spindle orientation variability of BPs upon wt LGN overexpression combined with minimal taxol incubation.
E13.5 wt forebrains were subjected to in utero electroporation of 6Myc-LGN-wt and 18 hr later (E14.25) incubated for 2.5 hr in BRO culture with either solvent (DMSO) only (Control) or with 25 pM taxol. (A, B) Live tissue imaging of spindle orientation as reported by chromosome plate orientation in organotypic slice culture of coronal sections from brains similar to those used in A, B, in control or 25 pM taxol incubations on the microscope stage. Electroporated cells were identified by expression of GAP43-EGFP co-electroporated with 6Myc-LGN-wt. Time-lapse is 2.5 min. 0 min corresponds to anaphase onset. Merge: chromosomes in red, EGFP in green. Vertical or oblique dashed lines indicate chromosome plate orientations in metaphase or anaphase onset; maximal deviation angles for each plate were set by the orientation at an early time-point (yellow) and a later time-point (red), which are quantified in F. White dashed lines indicate intermediate orientations. Scale bar = 5 μm. (C) Mean ± SEM of the maximal amplitude of deviations for electroporated BPs in control or taxol incubations. The mean number of apical/basal astral microtubules of BPs was not significantly different between control and taxol-incubated BPs; n = 8 progenitors per category, from 2 independent litters and experiments.
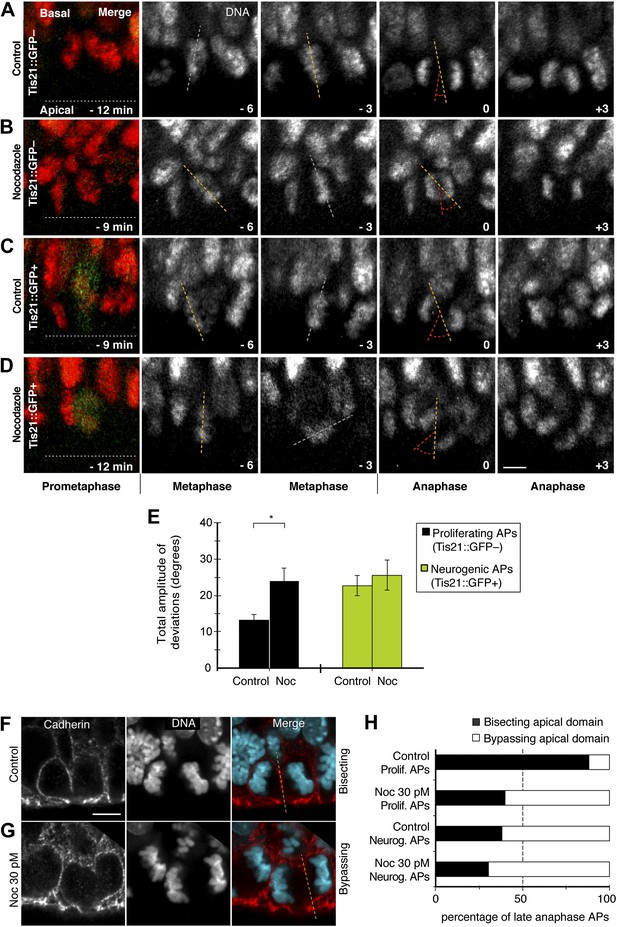
Dynamic spindle orientation variability and asymmetric AP divisions increase with minimal nocodazole.
E14.5 forebrains from Tis21::GFP mice were incubated on the microscope stage with either solvent (DMSO) only (Control) or with 30 pM nocodazole (Noc). (A–D) Live tissue imaging of spindle orientation, as reported by chromosome plate orientation, in organotypic slice culture of coronal sections from E14.5 Tis21::GFP mouse dorsolateral telencephalon. 0 min corresponds to anaphase onset. Apical progenitors underwent either proliferative (A, B, Tis21::GFP–) or neurogenic (C, D; Tis21::GFP+) divisions. Merge: chromosomes in red, EGFP in green. Chromosome plate orientation was determined by measuring angular deviations from the apico-basal axis (0°, corresponding to vertical), which runs perpendicular to the apical ventricular surface (90°, horizontal white dotted lines in A–D). Time-lapse is 3 min. Vertical or oblique dashed lines indicate chromosome plate orientations in metaphase or anaphase onset; maximal deviation angles for each plate were set by the orientation at an early time-point (yellow) and a later time-point (red), which are quantified in E. White dashed lines indicate intermediate orientations. Scale bar = 5 μm. (E) Mean ± SEM of the maximal amplitude of deviations for proliferating (Tis21::GFP–) vs neurogenic (Tis21::GFP+) APs, either in control or nocodazole incubations, *p<0.05, K–W with DMC post test, n = 20 progenitors per category, from 3 independent litters and experiments. See also Figure 7. (F, G) Cadherin immunofluorescence combined with DNA staining (DAPI) to reveal symmetric vs asymmetric divisions of anaphase APs with regard to the apical domain, as reported by bisecting or bypassing, respectively, of the cadherin-negative apical plasma membrane (dashed lines). Merge: cadherin, red; DNA, cyan. (H) Percentages of late anaphase APs, with advanced cytokinetic cleavage furrow, prospectively bisecting or bypassing their apical domain; n = 20 progenitors per category, from three independent litters and experiments. Scale bars = 5 μm.
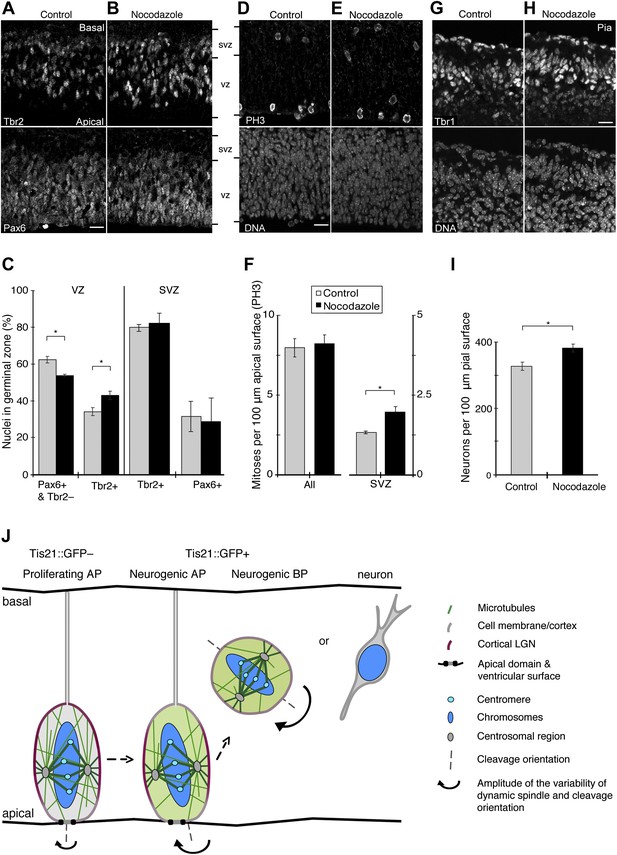
Increase in basal progenitors, basal mitoses, and neurogenesis with minimal nocodazole.
E13.5 forebrains from wt mice were incubated for 24 hr in BRO culture, either with solvent (DMSO) only (Control) or with 30 pM nocodazole, followed by immunofluorescence of coronal sections of the dorsolateral telencephalon, acquisition of 1 μm confocal sections (A, B, D, E, G, H) and quantification (C, F, I). (A, B) Tbr2 (top) and Pax6 (bottom) double staining. VZ, ventricular zone; SVZ, subventricular zone. (C) Left: percentages of nuclei, identified by DAPI staining (not shown), in the VZ that are either resident APs (Pax6-positive and Tbr2-negative) or newborn BPs (Tbr2-positive). Control vs nocodazole: APs, *p=0.002; BPs, *p=0.004. Right: percentages of nuclei in the SVZ that are positive for Tbr2 or Pax6. (D–E) Staining for the mitosis marker PH3 (top) and DNA (DAPI, bottom). (F) Number and location of mitotic cells per 100 μm of apical (ventricular) surface; All, sum of mitoses in VZ and SVZ. Control vs nocodazole: SVZ, *p=0.031. (G, H) Staining for the neuronal marker Tbr1 (top) and DNA (DAPI, bottom). Note that the Tbr1 immunoreactivity basal to the cortical plate has been reported before (Englund et al., 2005); it may also reflect tissue stretching during cryosectioning. (I) Number of neurons per 100 μm of pial surface. Control vs nocodazole: *p=0.015. (C, F, I) One-tail t-tests, n = 3 independent litters and experiments; error bars are SEM. (A, B, D, E, G, H) Scale bars = 20 μm. See also Figure 10—figure supplement 1. (J) Conceptual model of mitotic spindle orientation control by local astral microtubule abundance, in proliferating vs neurogenic progenitors. We propose that mitotic astral microtubules that reach the apical or basal cell cortex (light green rods) of neural stem and progenitor cells regulate the dynamics of spindle orientation variability. Like guy ropes do for a camping tent, these astrals help anchor the spindle to the cell cortex, most likely through interactions with factors that have specific cortical enrichments, such as LGN (red; colour intensity indicates local abundance in the three progenitor types shown). In APs undergoing proliferative division (Tis21::GFP–), these apical/basal astrals at relatively high abundance minimize deviations in spindle orientation, thereby maintaining the canonical cleavage orientation, perpendicular to the apical (ventricular) surface, and thus favouring symmetric proliferative divisions. In neurogenic (Tis21::GFP+) APs, and even more so in BPs, a reduction in the number of these astrals decreases cortical anchoring and increases random deviations in spindle orientation. For APs, this favours asymmetric neurogenic divisions that can generate a BP or a neuron. By contrast, astrals that reach the central cell cortex (medium green rods) of neural progenitors may be more involved in the fundamental establishment of a centrally located and functional bipolar spindle that can congress, and then faithfully segregate, chromosomes. (Dark green rods are kinetochore microtubules; for simplicity, other microtubule populations, such as interpolar microtubules, are omitted.)
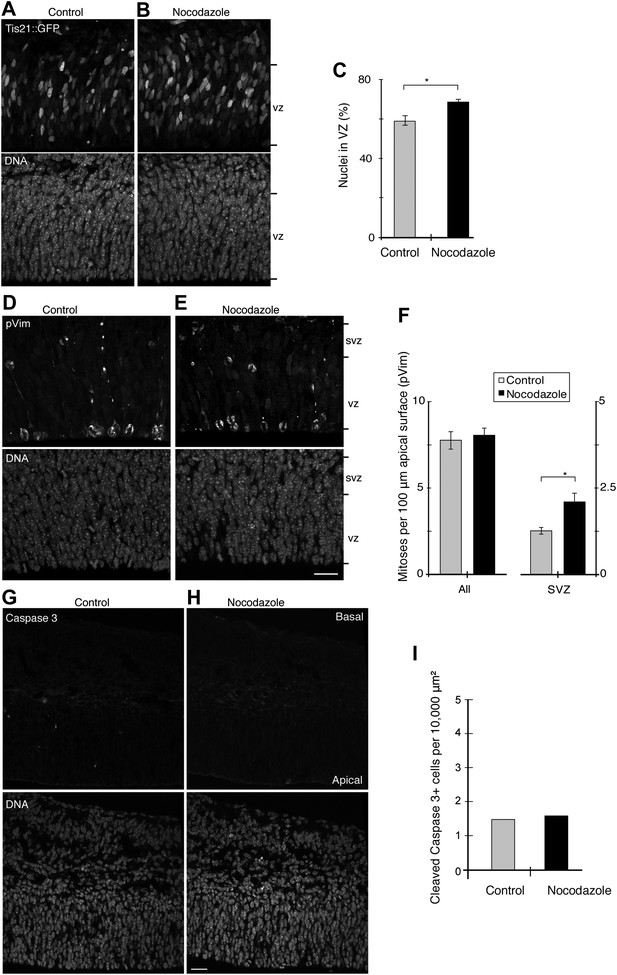
BRO culture with 30 pM nocodazole increases Tis21-positive VZ cells and basal mitoses, but not apoptosis.
E13.5 forebrains from wt mice were incubated for 24 hr in BRO culture, either with solvent (DMSO) only (control) or with 30 pM nocodazole, followed by immunofluorescence of coronal sections of the dorsolateral telencephalon, acquisition of 0.9 μm confocal sections (A, B, D, E, G, H) and quantification (C, F, I). (A, B) Tis21::GFP (top) and DNA (DAPI, bottom) double staining. (C) Percentages of nuclei (by DAPI) in the VZ that are Tis21::GFP-positive. Control vs nocodazole: *p=0.030. (D–E) Staining for the mitosis marker phosphovimentin (pVim, top) and DNA (bottom). (F) Number and location of mitotic cells per 100 μm of apical (ventricular) surface; All, sum of mitoses in VZ and SVZ. Control vs nocodazole: SVZ, *p=0.026; n = 3 brains from two independent litters and experiments. (G, H) Immunofluorescence for cleaved caspase 3 (top) and DAPI staining (bottom). (I) Pooled number of caspase 3-positive cells per 10,000 μm2 of dorsolateral cortex tissue; n = 3 brains from three independent litters and experiments. Error bars are SEM. Scale bars = 20 μm.
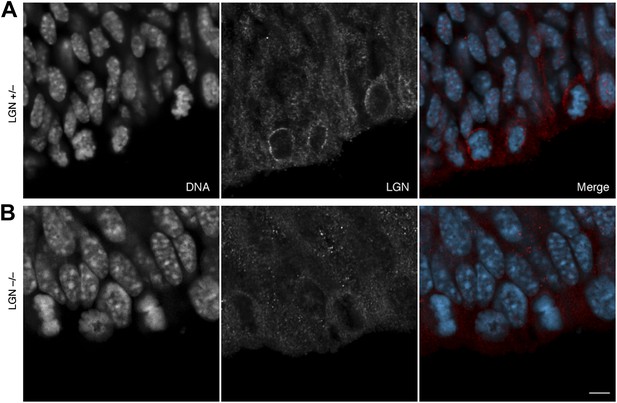
No LGN immunoreactivity in LGN –/– mice.
E13.5 LGN+/– (top) and LGN–/– (bottom) mice (as in Konno et al, 2008). 1-μm confocal sections of immunofluorescences for endogenous LGN (red), together with DNA (DAPI, cyan) staining, of coronal sections of the dorsolateral telencephalon. Scale bar = 5 μm.
Videos
Dynamic spindle orientation variability in a representative proliferating AP. Related to Figure 1A.
Live tissue imaging of spindle orientation, as reported by the chromosome plate (DNA) orientation, in organotypic slice culture of dorsolateral telencephalon coronal sections, from an E14.5 Tis21::GFP mouse. Time-lapse 3 min. Total time elapsed 18 min.
Dynamic spindle orientation variability in a representative neurogenic AP. Related to Figure 1B.
Live tissue imaging of spindle orientation, as reported by the chromosome plate (DNA) orientation, in organotypic slice culture of dorsolateral telencephalon coronal sections, from an E14.5 Tis21::GFP mouse. Time-lapse 3 min. Total time elapsed 18 min.
Dynamic spindle orientation variability in a representative neurogenic BP. Related to Figure 1C.
Live tissue imaging of spindle orientation, as reported by the chromosome plate (DNA) orientation, in organotypic slice culture of dorsolateral telencephalon coronal sections, from an E14.5 Tis21::GFP mouse. Time-lapse 3 min. Total time elapsed 18 min.
Dynamic spindle orientation variability in a representative proliferating AP incubated with DMSO only at 1.5 min time resolution. Related to Figure 8A.
Live tissue imaging of spindle orientation, as reported by the chromosome plate (DNA) orientation, in organotypic slice culture of dorsolateral telencephalon coronal sections, from an E14.5 Tis21::GFP mouse. Total time elapsed 15 min.
Dynamic spindle orientation variability in a representative proliferating AP incubated with 25 pM taxol at 1.5 min time resolution. Related to Figure 8B.
Live tissue imaging of spindle orientation, as reported by the chromosome plate (DNA) orientation, in organotypic slice culture of dorsolateral telencephalon coronal sections, from an E14.5 Tis21::GFP mouse. Total time elapsed 13.5 min.
Dynamic spindle orientation variability in a representative neurogenic AP incubated with DMSO only at 1.5 min time resolution.
Related to Figure 8C. Live tissue imaging of spindle orientation, as reported by the chromosome plate (DNA) orientation, in organotypic slice culture of dorsolateral telencephalon coronal sections, from an E14.5 Tis21::GFP mouse. Total time elapsed 10.5 min.
Dynamic spindle orientation variability in a representative neurogenic AP incubated with 25 pM taxol at 1.5 min time resolution.
Related to Figure 8D. Live tissue imaging of spindle orientation, as reported by the chromosome plate (DNA) orientation, in organotypic slice culture of dorsolateral telencephalon coronal sections, from an E14.5 Tis21::GFP mouse. Total time elapsed 12 min.
Dynamic spindle orientation variability in a representative AP incubated with DMSO only and co-electroporated with GAP43-EGFP and 6Myc-LGN-wt.
Related to Figure 8E. Live tissue imaging of spindle orientation, as reported by the chromosome plate (DNA) orientation, in organotypic slice culture of dorsolateral telencephalon coronal sections, from an E14.5 wt mouse. Time-lapse 3 min. Total time elapsed 17.5 min.
Dynamic spindle orientation variability in a representative AP incubated with 25 pM taxol and co-electroporated with GAP43-EGFP and 6Myc-LGN-wt.
Related to Figure 8F. Live tissue imaging of spindle orientation, as reported by the chromosome plate (DNA) orientation, in organotypic slice culture of dorsolateral telencephalon coronal sections, from an E14.5 wt mouse. Time-lapse 3 min. Total time elapsed 20.5 min.
Dynamic spindle orientation variability in a representative proliferating AP incubated with DMSO only.
Related to Figure 9A. Live tissue imaging of spindle orientation, as reported by the chromosome plate (DNA) orientation, in organotypic slice culture of dorsolateral telencephalon coronal sections, from an E14.5 Tis21::GFP mouse. Time-lapse 3 min. Total time elapsed 15 min.
Dynamic spindle orientation variability in a representative proliferating AP incubated with DMSO only.
Related to Figure 9B. Live tissue imaging of spindle orientation, as reported by the chromosome plate (DNA) orientation, in organotypic slice culture of dorsolateral telencephalon coronal sections, from an E14.5 Tis21::GFP mouse. Time-lapse 3 min. Total time elapsed 12 min.
Dynamic spindle orientation variability in a representative neurogenic AP incubated with 30 pM nocodazole.
Related to Figure 9C. Live tissue imaging of spindle orientation, as reported by the chromosome plate (DNA) orientation, in organotypic slice culture of dorsolateral telencephalon coronal sections, from an E14.5 Tis21::GFP mouse. Time-lapse 3 min. Total time elapsed 12 min.
Dynamic spindle orientation variability in a representative neurogenic AP incubated with 30 pM nocodazole.
Related to Figure 9D. Live tissue imaging of spindle orientation, as reported by the chromosome plate (DNA) orientation, in organotypic slice culture of dorsolateral telencephalon coronal sections, from an E14.5 Tis21::GFP mouse. Time-lapse 3 min. Total time elapsed 15 min.





