A single vertebrate DNA virus protein disarms invertebrate immunity to RNA virus infection
Figures
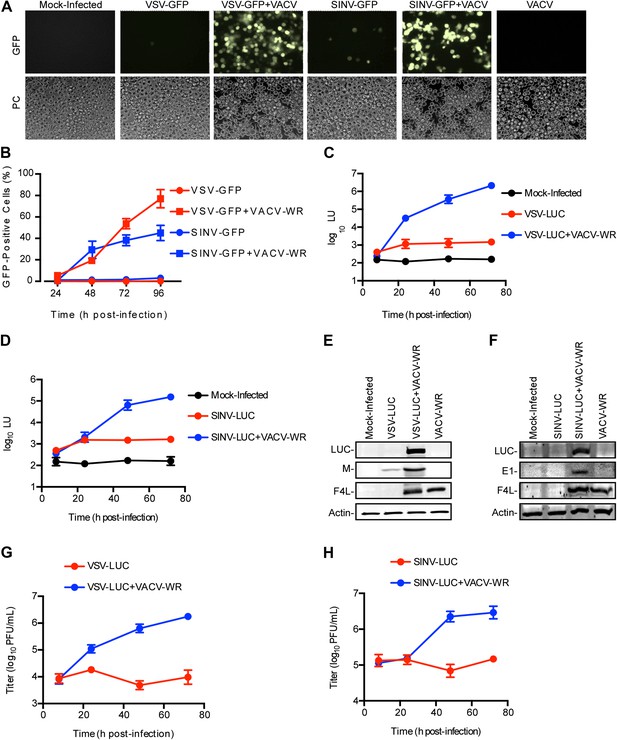
Restriction of RNA virus replication in LD652 cells is relieved by VACV co-infection.
(A) GFP fluorescence (top) and phase contrast images (PC, bottom) of infected LD652 cells at 96 hpi. Images are shown at 20X magnification. (B) Percentage of GFP-positive LD652 cells from experiments in (A). (C and D) LUC assay [arbitrary light units (LU)] of lysates from mock-infected cells or cells infected with VSV-LUC (C) or SINV-LUC (D), in the absence or presence of VACV-WR. Mock-infected data are identical in (C) and (D). (E and F) Immunoblot of LUC, VSV M (E) or SINV E1 (F), VACV F4L, and cellular actin proteins in lysates from (C) and (D) 72 hpi. (G and H) VSV-LUC (G) or SINV-LUC (H) titers, (plaque-forming units (PFU)/ml) in culture supernatants from (C) and (D), respectively. Quantitative data represent means (±SEM). See also Figure 1—figure supplement 1.
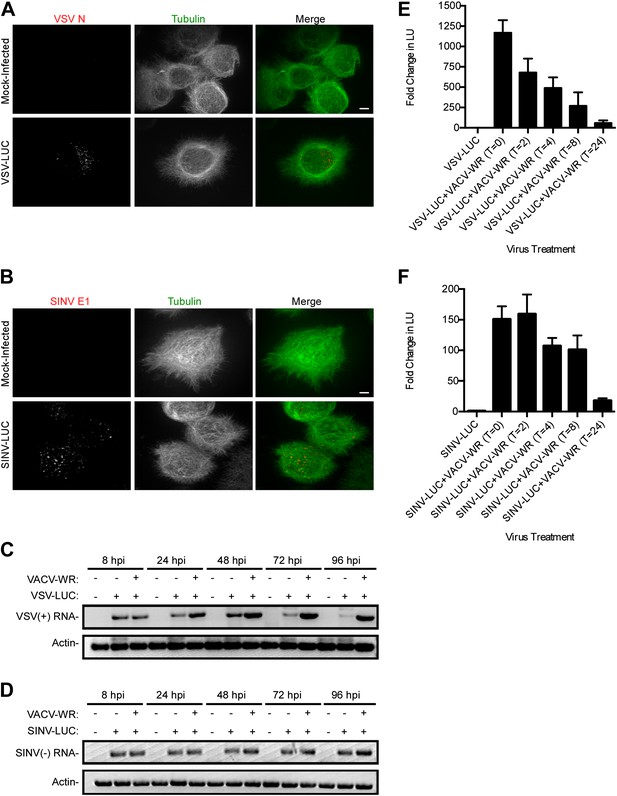
Restriction of RNA virus replication in L. dispar cells occurs at a step post-entry.
VSV N (A) and SINV E1 (B) virion protein staining inside LD652 cells by confocal microscopy 24 hpi. Scale bars represent 10 μm. (C) RT-PCR assay of VSV (+)-sense transcription and cellular actin mRNA levels in LD652 cells at the indicated times post-infection. (D) RT-PCR assay of SINV (−)-sense transcription and cellular actin mRNA levels in LD652 cells at the indicated time post-infection. (E and F) Effect of timing of VACV-WR co-infection on VSV-LUC (E) or SINV-LUC (F) gene expression. Cells were first infected with the indicated RNA virus and then infected with VACV immediately (T = 0) or at the indicated hours (T = 2–24) post-RNA virus infection. Lysates were collected for LUC assay (shown as arbitrary light units [LU]) 72 hr post RNA virus infection. Fold change in LU was calculated by dividing LU obtained from each co-infection condition by LU obtained from cells infected with VSV-LUC or SINV-LUC in the absence of VACV-WR. Data represent means (+SEM).
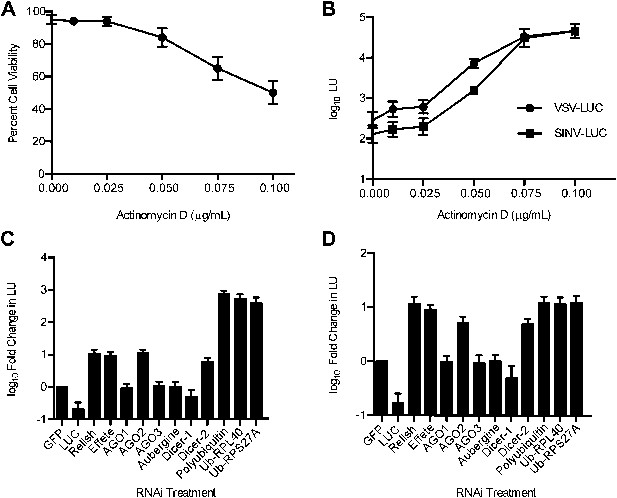
Host transcription and RNAi-, NF-κB-, and ubiquitin-related factors restrict RNA virus replication in LD652 cells.
(A) Trypan blue exclusion assay to measure cell viability in the presence of ActD (48 hr). (B) Effect of ActD on virus LUC activity (in LU) 48 hpi. (C and D) LUC activity (LU) in lysates from cells 48 hpi with either VSV-LUC (C) or SINV-LUC (D) and after RNAi of L. dispar transcripts relative to LU generated in GFP (control) RNAi treatments. Data represent means (+SEM). See also Figure 2—figure supplement 1.
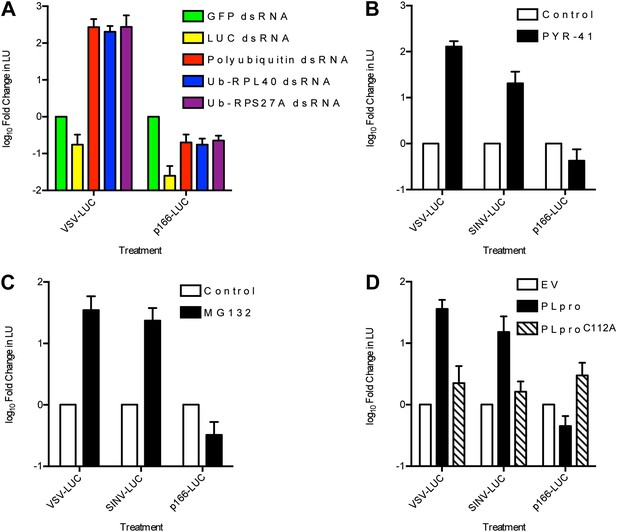
RNA virus gene expression is enhanced upon inhibition of the ubiquitin-proteasome system in L. dispar cells.
(A) RNAi knockdown of ubiquitin (Ub) transcripts increases viral gene expression. LD652 cells were transfected with the indicated dsRNAs for 24 hr and then infected with the indicated viruses for 48 hr. In parallel experiments, uninfected cells were transfected with a p166 vector encoding LUC (p166-LUC) along with each indicated dsRNA. Lysates were collected 72 hr post-transfection and analyzed for LUC activity (in LU). Data are plotted as fold change when compared to LU generated in GFP dsRNA (control) treatments within each treatment group. (B and C) Effect of the E1 Ub-activating enzyme inhibitor PYR-41 (B) or the proteasome inhibitor MG132 (C) on virus gene expression. Cells were infected with the indicated viruses or transfected with p166-LUC. Media was replaced after 2 hr of infection (VSV/SINV-LUC) or 5 hr of transfection (p166-LUC) with either vehicle-containing medium (control) or medium containing 50 µM PYR-41 or 40 µM MG132. Lysates were prepared 48 hr later and assessed for LUC activity. Data are plotted as fold change in LU when compared to LU generated in control medium treatments. (D) Effect of expression of coronavirus PLpro deubiquitinase on virus gene expression. LD652 cells were transfected with empty pIZ/His-V5 vector (EV) or vectors encoding PLpro or the catalytic mutant form PLproC112A for 24 hr followed by infection with the indicated viruses. In parallel cultures, uninfected cells were co-transfected with the indicated pIZ/His-V5 constructs along with p166-LUC. In each case, cells were collected 48 hpi/transfection and analyzed for LUC activity. Data are plotted as fold change in LU when compared to LU generated in empty vector transfection treatments. Data represent means (+SEM).
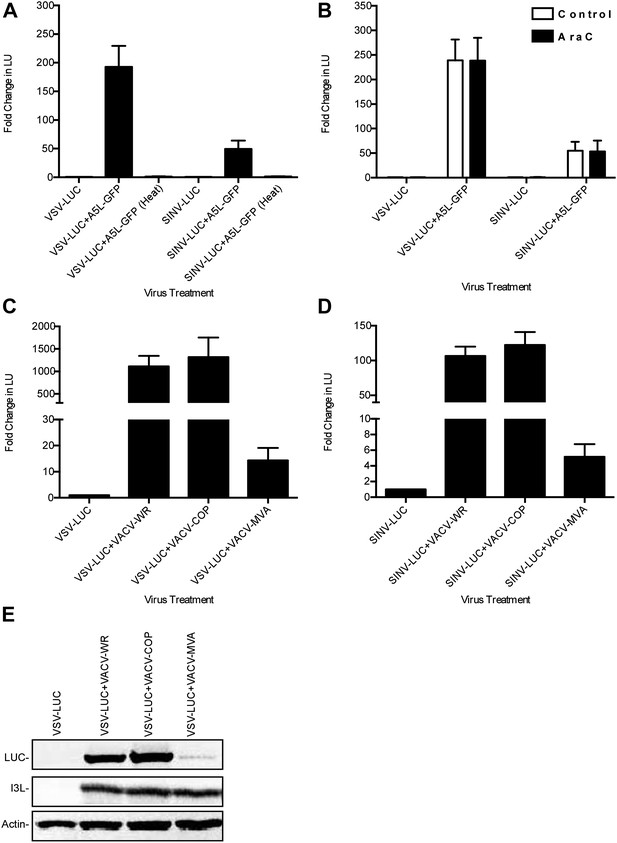
Characterization of VACV-dependent RNA virus rescue in LD652 cells.
(A) Effect of heat inactivation of VACV strain A5L-GFP on RNA virus rescue 48 hpi by LUC assay (in LU). LU generated from co-infection lysates and are plotted as 'fold change' with respect to single infection conditions. (B) Effect of AraC (200 μg/ml) treatment on A5L-GFP-mediated RNA virus rescue 48 hpi. Fold change in LU was calculated as in (A). (C and D) Relative LUC activity in lysates from cells infected with VSV-LUC (C) or SINV-LUC (D) and co-infected with various VACV strains and in the presence of AraC for 72 hr. Fold change in LU was calculated as in (A) VACV strain Copenhagen (VACV-COP); modified VACV Ankara (VACV-MVA). (E) Immunoblot of lysates from (C) for LUC, I3L, and actin. Data represent means (+SEM).
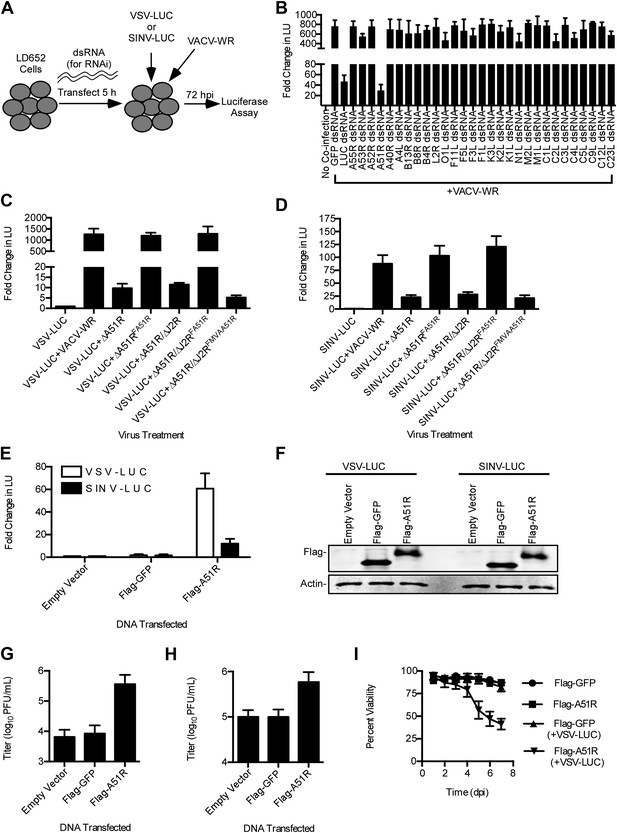
A51R is a major determinant of VACV-mediated rescue of RNA virus replication in LD652 cells.
(A) Strategy to identify VACV 'rescue' factors using dsRNA-mediated RNAi in L. dispar cells. (B) Relative LUC activity 72 hpi in lysates from cells infected with VSV-LUC and VACV-WR after the indicated RNAi treatment. Fold change in LU was calculated as in Figure 3A. (C and D) Relative LUC activity in lysates from cells co-infected with recombinant VACV strains and VSV-LUC (C) or SINV-LUC (D) in the presence of AraC (200 μg/ml) for 72 hr. (E) Relative LUC activity in lysates of cells transfected with empty p166 vector, Flag-GFP, or Flag-A51R for 24 hr and then infected with VSV-LUC or SINV-LUC for 48 hr. Fold change in LU are relative to LU obtained from empty vector treatment. (F) Immunoblot of Flag-GFP (lower band) and Flag-A51R (upper band) from lysates in (E). Actin served as a loading control. (G–H) VSV-LUC (G) and SINV-LUC (H) titers from experiments in (E). (I) Cell viability after VSV infection as measured by trypan blue exclusion assay. Cells were transfected for 24 hr with either Flag-GFP or Flag-A51R vectors and then mock-infected or infected with VSV-LUC. Data represent means (+SEM). See also Figure 4—figure supplements 1 and 2.
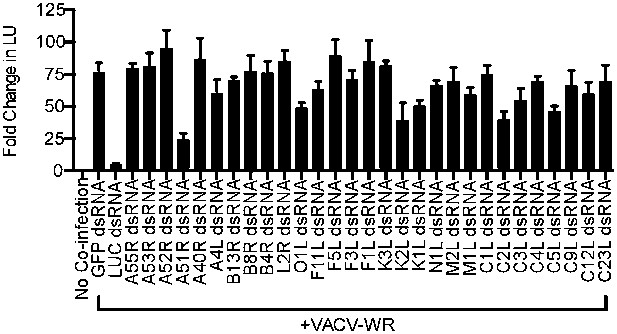
Identification of VACV A51R as a determinant of SINV rescue in L. dispar cells.
RNA interference (RNAi) assay in LD652 cells to identify VACV-encoded determinants of SINV rescue. L. dispar cells were treated as described in the legend for Figure 4A,B and then analyzed by LUC assay. Data represent means (+SEM).
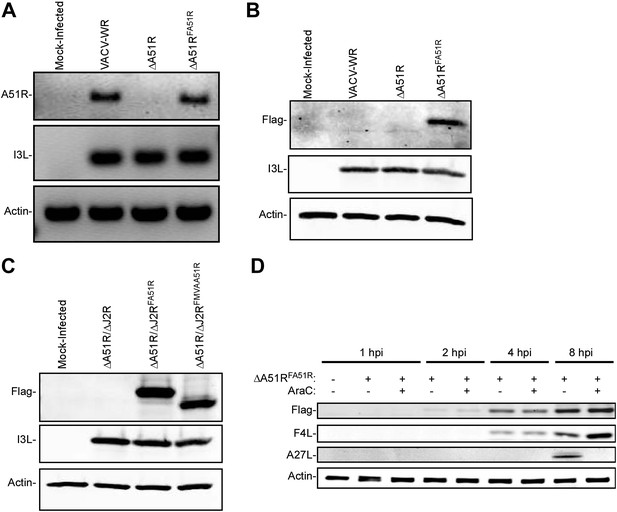
Characterization of A51R-related recombinant VACV strains.
(A) Agarose gel showing RT-PCR analysis of A51R, I3L, and actin mRNA expression in recombinant VACV strains 24 hpi in LD652 cells. (B) Immunoblot of cell lysates from BSC-40 cells infected for 24 hr with the indicated strains (MOI = 1) for Flag-A51R, VACV I3L and actin. (C) Immunoblot of BSC-40 cell lysates infected with the indicated recombinant VACV strains (MOI = 1) for 24 hr. Lysates were probed with antibodies for Flag (to detect Flag-A51R/Flag-MVAA51R), I3L, and actin. (D) Immunoblot of Flag-A51R protein expression during ΔA51RFREV infection (MOI = 1) of BSC-40 cells. Lysates were prepared at the indicated times post-infection and analyzed by immunoblot for: early VACV proteins (F4L), late VACV proteins (A27L), Flag (Flag-A51R), and actin. Where indicated, 50 μg/ml AraC was present in the cell culture medium throughout the time course.
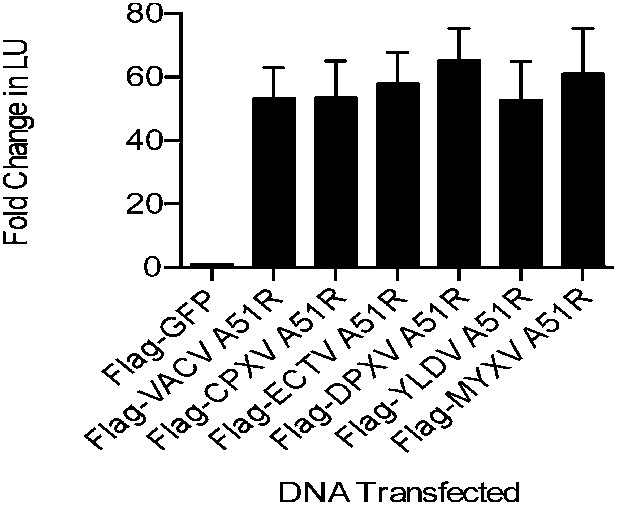
A51R proteins from disparate poxviruses rescue RNA virus replication in LD652 cells.
Relative LUC activity in lysates of cells expressing the indicated A51R construct and infected with VSV-LUC for 48 hr. Fold change in LU are relative to LU obtained in Flag-GFP treatments. CPXV, cowpox virus; ECTV, ectromelia virus; DPXV, deerpox virus; YLDV, Yaba-like disease virus; and MYXV, myxoma virus. Data represent means (+SEM).
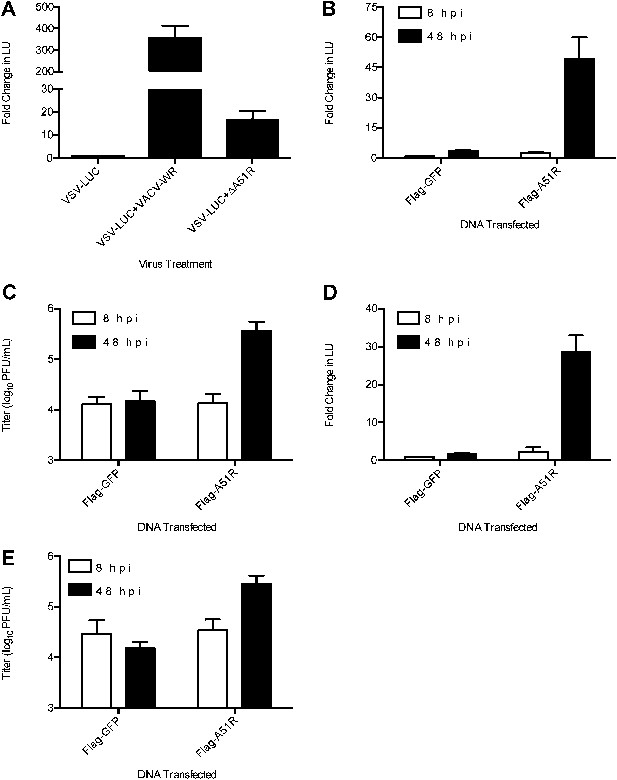
A51R relieves RNA virus restriction in multiple Lepidopteran hosts.
(A) Relative LUC activity in lysates of L. dispar-derived embryonic (LdEP) cells co-infected with VSV-LUC and VACV-WR or ΔA51R strains. Fold changes in LU are relative to LU obtained from VSV-LUC (8 hpi) lysates. (B) Relative LUC activity in lysates of Spodoptera frugiperda-derived Sf9 cells expressing Flag-GFP or Flag-A51R and infected with VSV-LUC. Fold change in LU are relative to LU obtained from Flag-GFP (8 hpi) lysates. (C) Virus titer of supernatants from (B). (D) Relative LUC activity in lysates of Manduca sexta-derived GV-1 cells expressing Flag-GFP or Flag-A51R and infected with VSV-LUC. Fold change in LU were calculated as in (B). (E) Virus titer of supernatants from (D). Data represent means (+SEM). See also Figure 6—figure supplement 1.

Effect of Flag-A51R expression or VACV-WR co-infection on VSV-LUC gene expression in Drosophila cells.
(A) Effect of Flag-A51R expression on VSV-LUC gene expression in DL1 cells. Cells were transfected with p166 vectors encoding Flag-A51R or Flag-GFP and infected with VSV-LUC (MOI = 0.1). 24 hpi, lysates were analyzed by LUC assay. Fold change in LU was calculated by dividing LU in Flag-A51R treatments with LU in Flag-GFP treatments. (B) Immunoblot of lysates from experiments in (A) for Flag-GFP (lower band), Flag-A51R (upper band), and actin. (C) Effect of VACV-WR co-infection on LUC activity. DL1 cells were infected with VSV-LUC (at the indicated MOIs) in the absence or presence of VACV-WR (MOI = 10). At the indicated hpi, lysates were analyzed by LUC assay. Data represent means (±SEM). (D) Immunoblot of lysates in (C) where cells were mock-infected or infected with VSV-LUC (MOI = 0.01) in the absence or presence of VACV-WR (MOI = 10) for 48 hr. Lysates were blotted for VSV M, VACV I3L, and actin proteins. Data represent means (+SEM).
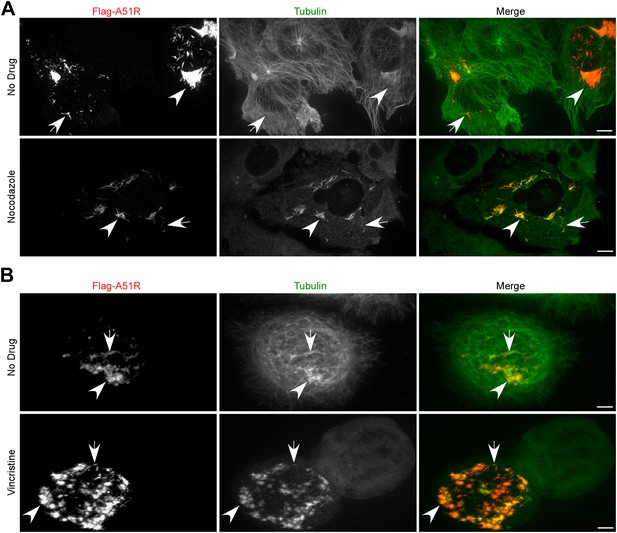
Poxvirus A51R proteins localize with and stabilize host microtubules.
(A) Immunofluorescence of Flag-A51R (red) and tubulin (green) in BSC-40 cells in the presence or absence of nocodazole 24 hr post-transfection of Flag-A51R vector. (B) Immunofluorescence of Flag-A51R (red) and tubulin (green) in LD652 cells in the presence or absence of vincristine 24 hr post-transfection of Flag-A51R vector. In (A) and (B) arrows indicate A51R filaments and arrowheads indicate A51R aggregate structures that overlap with tubulin staining. Note the absence of these aggregates and filaments in cells that lack Flag-A51R staining. Scale bars represent 10 μm. See also Figure 7—figure supplements 1 and 2.
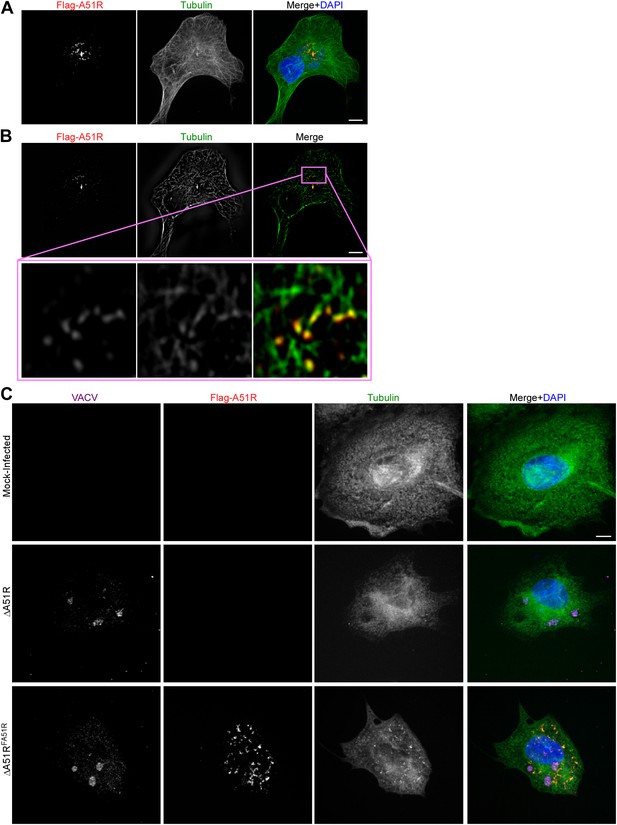
A51R localizes with and stabilizes MTs.
(A) Flag-A51R localization during VACV infection of BSC-40 cells. Cells were infected with the ΔA51RFREV strain (MOI = 3) and methanol fixed 8 hpi for confocal microscopy analysis using antibodies against Flag (red) and tubulin (green). DAPI staining (blue) was also used to mark nuclear and viral DNA in the merged image. (B) A single slice from the Z-stack shown in (A) illustrating Flag-A51R staining (red) colocalizing with tubulin staining (green). The inset shows a magnified view of Flag-A51R staining overlapping with MT tracks. (C) Nocodazole-resistant, MT-like filamentous tubulin pieces are only observed in VACV-infected cells when Flag-A51R is also present. BSC-40 cells were either mock-infected or infected with the indicated VACV strains (MOI = 3) for 8 hr. Coverslips were then methanol-fixed and processed for confocal microscopy using Flag (red), VACV (magenta), and tubulin (green) antibodies. DAPI staining (blue) was also used to mark DNA in merged images. Cell culture media contained 20 μM nocodazole from 2-8 hpi.
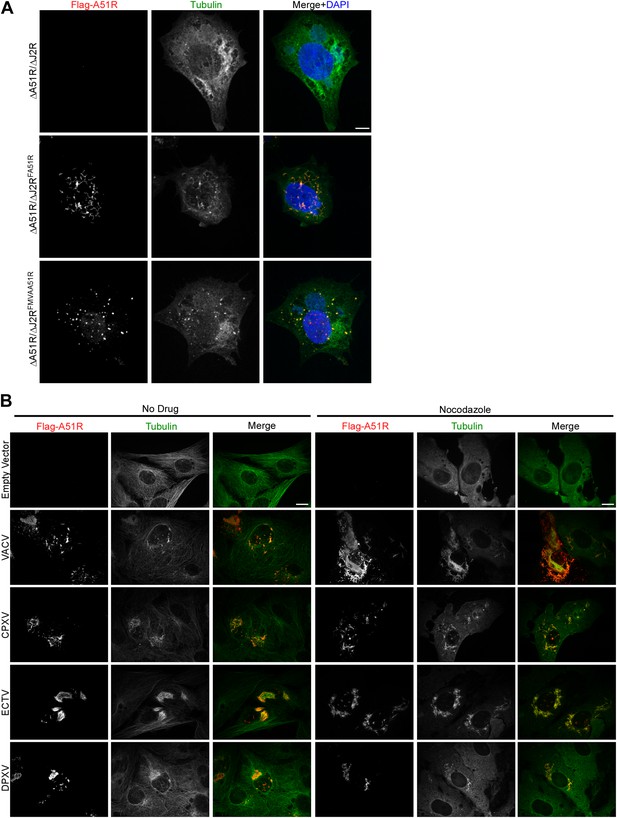
Localization of Flag-A51R proteins encoded by VACV-MVA and disparate poxviruses in BSC-40 cells.
(A) Localization of Flag-A51R and Flag-MVAA51R proteins in the presence of nocodazole (20 μM). Cells were infected with the indicated strains (MOI = 3) and then paraformaldehyde-fixed 8 hpi. Cells were stained with antibodies against Flag (red) and tubulin (green). DAPI staining (blue) was included in merge images. (B) Localization of Flag-tagged A51R proteins from the indicated poxviruses 24 hr post-transfection of BSC-40 cells with pCDNA3 expression plasmids. Where indicated, nocodazole (20 μM) was present in culture media from 5-24 hr post-transfection. Cells were then methanol-fixed and stained with Flag (red) and tubulin (green) antibodies. Scale bars represent 10 μm in each figure.
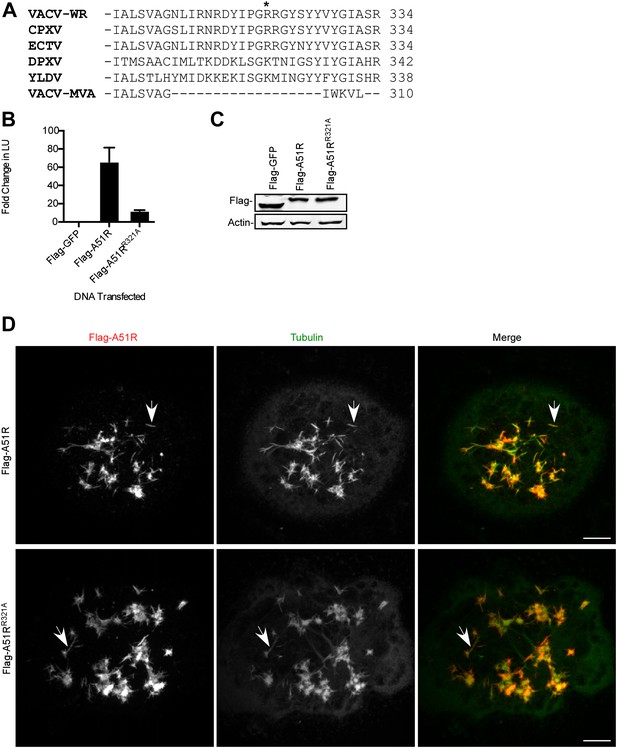
Microtubule stabilization is insufficient for full RNA virus rescue in LD652 cells.
(A) C-terminal alignment of poxvirus A51R proteins with site of R321A substitution indicated by an asterisk. Multiple alignments were performed using eBioX Software (v. 1.5.1) using T-COFFEE alignment parameters. CPXV, cowpox virus; ECTV, ectromelia virus; DPXV, deerpox virus; and YLDV, Yaba-like disease virus. (B) Relative LUC activity in lysates of cells transfected with Flag-GFP, Flag-A51R or Flag-A51RR321A p166 constructs for 24 hr and then infected with VSV-LUC for 48 hr. Fold change in LU are relative to LU obtained in Flag-GFP treatments. Data represent means (+SEM). (C) Immunoblot of Flag-GFP (lower band) and Flag-A51R/A51RR321A (upper bands) from lysates in (B). (D) Immunofluorescence of Flag-A51R/A51RR321A (red) and tubulin (green) proteins in LD652 cells in the presence of vincristine 48 hr post-transfection of Flag-A51R vectors. Arrows indicate A51R filaments that overlap with tubulin staining. Scale bars represent 10 μm.
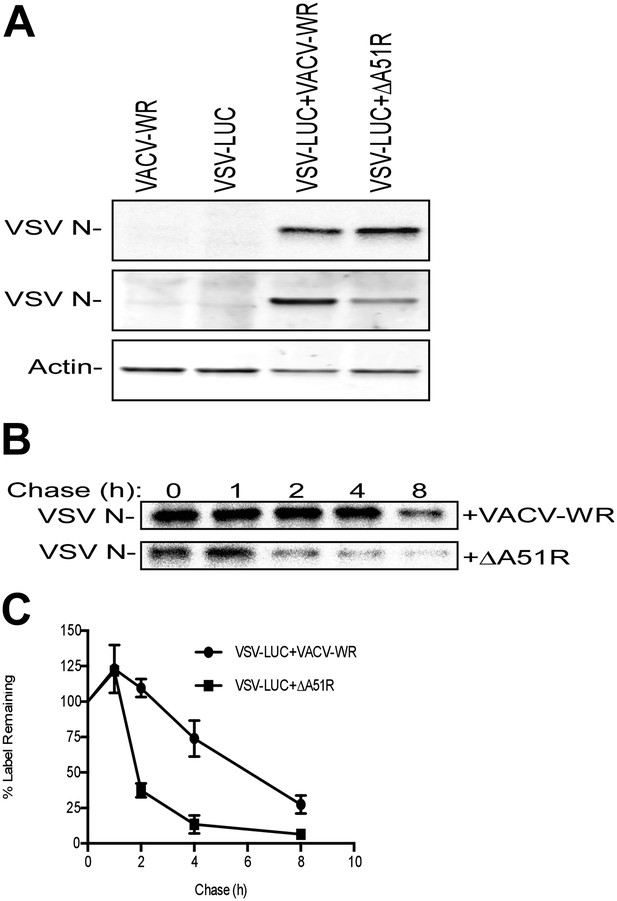
A51R promotes RNA virus protein stability in LD652 cells.
Cells were infected with the indicated strains for 72 hr and then pulsed for 2 hr with [35S]methionine. To visualize radiolabeled, nascent VSV N protein, cell lysates were subjected to immunoprecpitation with anti-VSV N protein antibodies and immunoprecipitated complexes were separated by SDS-PAGE and visualized by autoradiography (top panel). Equal fractions of each total cellular lysate were also used in parallel immunoblot experiments to detect total VSV N (middle panel) or actin protein levels. (B and C) Cells were infected with VSV-LUC and the indicated VACV strains as in (A), pulsed for 2 hr with [35S]methionine and then chased for the indicated times at which point cell lysates were collected and subjected to immunoprecipitation with anti-VSV N antibodies. Immunoprecipitated complexes were then separated by SDS-PAGE, visualized by autoradiography (B) and the percentage of radiolabeled VSV N protein remaining (compared to T = 0, representing the end point of the 2 hr pulse) at each time point post-pulse was plotted (C). Note that lysates containing radiolabeled VSV N protein from VACV-WR and ΔA51R co-infections time courses (B) were processed separately and thus band intensities between VACV infection treatments are not directly comparable. Data in (C) represent means (+SD).
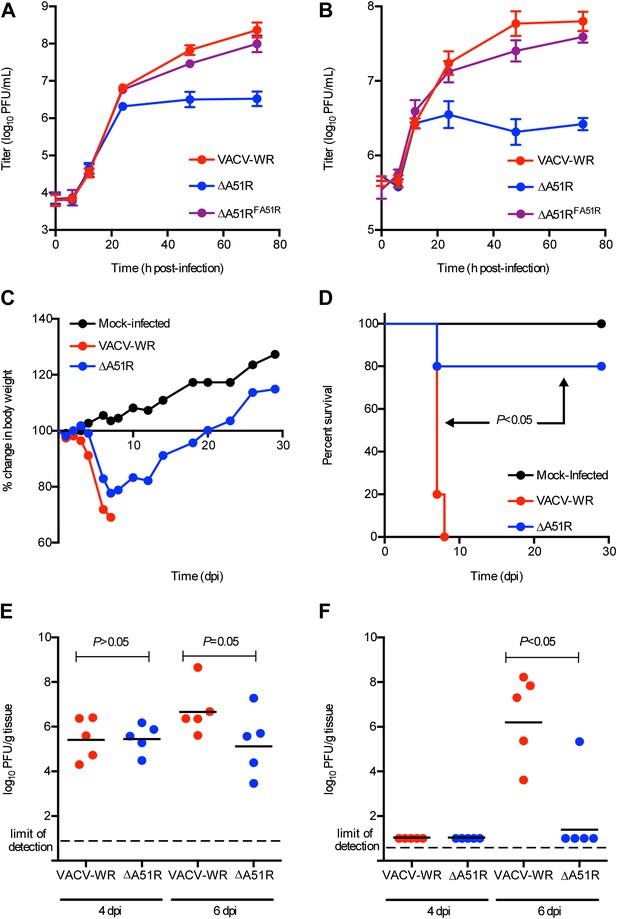
A51R promotes VACV replication and pathogenesis in vertebrates.
(A and B) VACV titers from BSC-40 cells infected with the indicated strains at low MOI (0.03) (A) or high MOI (3) (B). Data represent means (+/-SEM). (C) Body weight of NMRI mice infected with the indicated virus (10,000 PFU/animal). Data represent mean percent body weight change among surviving members in each group at the indicated day post-infection. (D) Percent survival of mice from (C). p<0.05 indicates a statistically significant difference in survival between VACV-WR- and ΔA51R strain-infected animals. (E and F) Virus titer in lung (F) and ovary (F) tissue from mice infected with VACV-WR or ΔA51R. Each dot indicates the total virus titers from an individual mouse. Horizontal bars represent the mean of each group. Mice were infected as in (C) and euthanized at 4 or 6 days post-infection. p<0.05 indicates a statistically significant difference between infection group tissue titers. See also Figure 10—figure supplement 1.
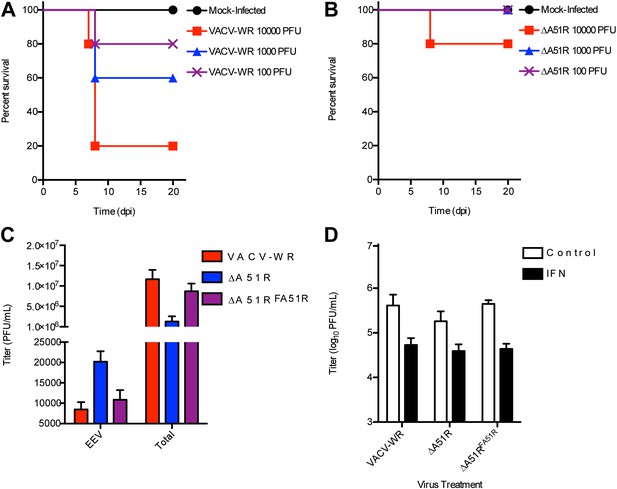
The ΔA51R strain displays attenuated pathogenesis in NMRI mice, releases increased levels of EEV, and does not display enhanced sensitivity to IFN treatment.
Groups of 5 NMRI mice were intranasally-inoculated with the indicated doses of either VACV-WR (A) or ΔA51R (B) strains and percentage survival of these groups of mice was tracked over the indicated time course. (C) Effect of A51R deletion on extracellular enveloped virus (EEV) production. BSC-40 cells were infected with the indicated strains at a MOI of 3 for 24 hr at which point supernatants were collected by low speed centrifugation, neutralized for intracellular mature virus (IMV) infectivity by incubation with L1R antibody (Backes et al., 2012) and titered on BSC-40 cell monolayers to determine ‘EEV’ titers. In parallel cultures ‘total’ virus titer (IMV+EEV) was determined by collecting both supernatants and cells, followed by three rounds of freeze-thaw and titration on BSC-40 cell monolayers. Data represent means (+SEM). (D) Effect of interferon (IFN) pretreatment on ΔA51R virus replication in BSC-40 cells. Cell culture medium containing 1000 U/ml recombinant IFN-α or normal growth medium (control) was used to replace growth medium of cells 16 hr prior to infection with the indicated viruses (MOI = 0.03). After 24 of infection, total virus titers were determined by plaque assay as in (C). Data represent means (+SEM).
Videos
Three-dimensional rendering of Flag-A51R-transfected LD652 cell in the presence of vincristine 48 hr post-transfection.
A merge between Flag (red) and tubulin (green) staining is shown.
Three-dimensional rendering of Flag-A51RR321A-transfected LD652 cell in the presence of vincristine 48 hr post-transfection.
A merge between Flag (red) and tubulin (green) staining is shown.
Tables
Differential conservation of Chordopoxirinae A51R genes
| Genus | A51R | Example species (% amino acid identity to VACV-WR A51R) |
|---|---|---|
| Orthopoxvirus | + | VACV* |
| HSPV† (96) | ||
| CPXV (94) | ||
| ECTV (93) | ||
| VARV (92) | ||
| Suipoxvirus | + | SPXV (33) |
| Yatapoxvirus | + | TANV (35) |
| YLDV (35) | ||
| Leporipoxvirus | + | MYXV (35) |
| SFV (35) | ||
| Capripoxvirus | + | GTPV (32) |
| SPPV (30) | ||
| LSDV (29) | ||
| Cervidpoxvirus | + | DPV (31) |
| Parapoxvirus | + | ORFV (22) |
| Molluscipoxvirus | – | MCV |
| Avipoxvirus | – | FPV |
| CNPV | ||
| Unclassified | – | CRV |
-
*
VACV strains MVA (Antoine et al., 1998) and Dryvax (Qin et al., 2011) contain a truncated and fragmented A51R gene, respectively.
-
†
HSPV contains a fragmented A51R gene (Tulman et al., 2006).
-
‘+’ Indicates presence, and ‘−‘ indicates absence of A51R gene in viral genomes within each genus. Abbreviations: VACV, vaccinia virus; HSPV, horsepox virus; TATV, taterapox virus; VARV, variola virus; SPXV, swinepox virus; TANV, tanapox virus; yaba-like disease virus; MYXV, myxoma virus; SFV, Shope fibroma virus; GTPV, goatpox virus; SPPV, sheeppox virus; LSDV, lumpy skin disease virus; DPV, deerpox virus; FPV, fowlpox virus; CNPV, canarypox virus; MCV, molluscum contagiosum; ORFV, orf virus; CRV, crocodilepox virus.
Proteins most-enriched in Flag-A51R immunoprecipitates from LD652 cells
| Protein | No. of spectral counts | ΔNSAF (NSAFFlag-A51R-NSAFControl) |
|---|---|---|
| Myosin II essential light chain | 110 | 0.0355 |
| Ub | 21 | 0.0113 |
| Flag-A51R | 33 | 0.0066 |
| Ribosomal protein S15 | 18 | 0.0056 |
Additional files
-
Supplementary file 1
L. dispar transcript sequences identified by mRNA-seq and primers used for dsRNA-mediated RNAi of L. dispar transcripts.
- https://doi.org/10.7554/eLife.02910.025
-
Supplementary file 2
Primers used for dsRNA-mediated RNAi of VACV transcripts.
- https://doi.org/10.7554/eLife.02910.026





