MicroRNA-mediated repression of nonsense mRNAs
Figures
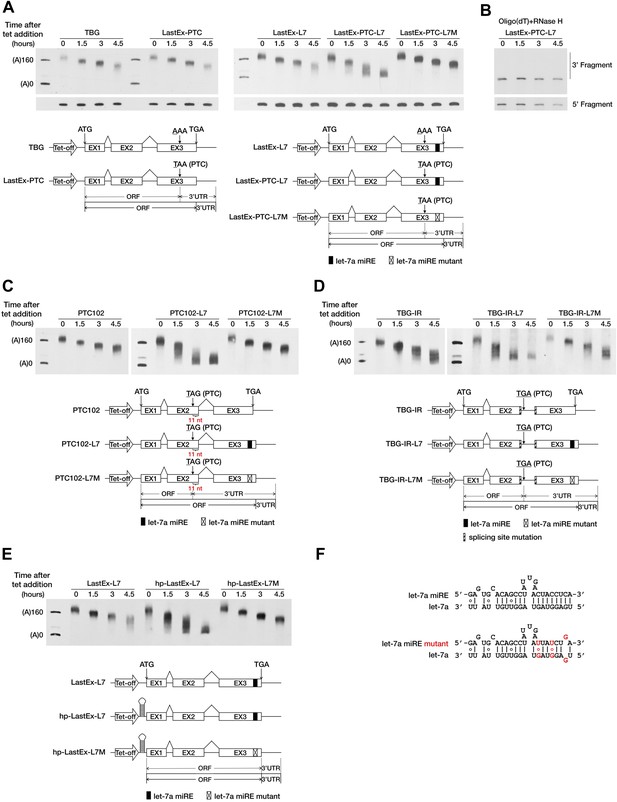
A PTC potentiates rapid miRNA-mediated deadenylation of nonsense mRNAs.
(A) The influence of a PTC on BG mRNAs with or without a downstream miRE. Cytoplasmic RNA was collected at the indicated times after transcriptional arrest by adding tetracycline (tet). The RNA samples were then treated with RNase H and an oligodeoxynucleotide complementary to codons 74–81 of BG mRNA to generate 5′ and 3′ fragments, which were then separated by electrophoresis on a denaturing polyacrylamide gel and detected by Northern blotting. Left panel: BG mRNA deadenylation with or without a PTC in the last exon. TBG contains an intact BG ORF. A PTC was introduced into TBG at codon 121 within the last exon to generate LastEx-PTC. Both constructs harbor no miRE in their ORFs. Right panel: the influence of let-7a on the deadenylation rate of BG mRNAs harboring one let-7a miRE in its ORF with or without an upstream PTC mutation. LastEx-L7 contains one let-7a miRE in the last exon of the BG ORF. LastEx-PTC-L7 has a PTC at the same position as LastEx-PTC, which is 16 nt upstream of the let-7a miRE. Two nucleotides within LastEx-PTC-L7 let-7a miRE seed were changed to create a synonymous codon in LastEx-PTC-L7M. The positions of the ORF start site and original stop codon are indicated by arrows. The borders of the original ORF/3′ UTR and redefined ORF/3′ UTR upon PTC mutation are indicated by solid or dashed lines below the constructs. Markers A(0) and A(160) correspond in size to BG mRNA 3′ fragments bearing no poly(A) or a 160-nt poly(A) tail, respectively. (B) Confirmation of poly(A) tail shortening by treatment with oligo(dT) and RNase H. The same LastEx-PTC-L7 RNA as in A were further treated with oligo(dT) and RNase H and analyzed by Northern blotting. (C) Induction by let-7a of accelerated deadenylation of nonsense BG mRNAs that do not conform to the ‘50 nt boundary rule’ of EJC-NMD. PTC102-L7 contains a PTC mutation 11 nt upstream of the last exon–exon junction and a let-7a miRE in the last exon of the ORF. PTC102-L7M is identical to PTC102-L7 except for a mutated let-7a miRE. PTC102 contains the PTC only. (D) The influence of let-7a on the deadenylation rate of BG mRNAs harboring one let-7a miRE in its ORF with a retained last intron. TBG-IR-L7 contains one let-7a miRE in the last exon of the BG ORF and a retained last intron that creates a PTC 505 nt upstream of the let-7a miRE. Two nucleotides within the TBG-IR-L7 let-7a miRE seed were mutated as in A. TBG-IR only has the retained intron but no miRE. (E) The influence of let-7a on the deadenylation rate of BG mRNAs harboring one let-7a miRE in its ORF in the absence of translation. hp-LastEx-L7 contains a 40-nt inverted repeat in its 5′ UTR to block translation initiation. hp-LastEx-L7M is identical to hp-LastEx-L7 except for a mutated let-7a miRE. (F) Duplexes expected for the let-7a miRE or its mutant counterpart base-paired with let-7a.
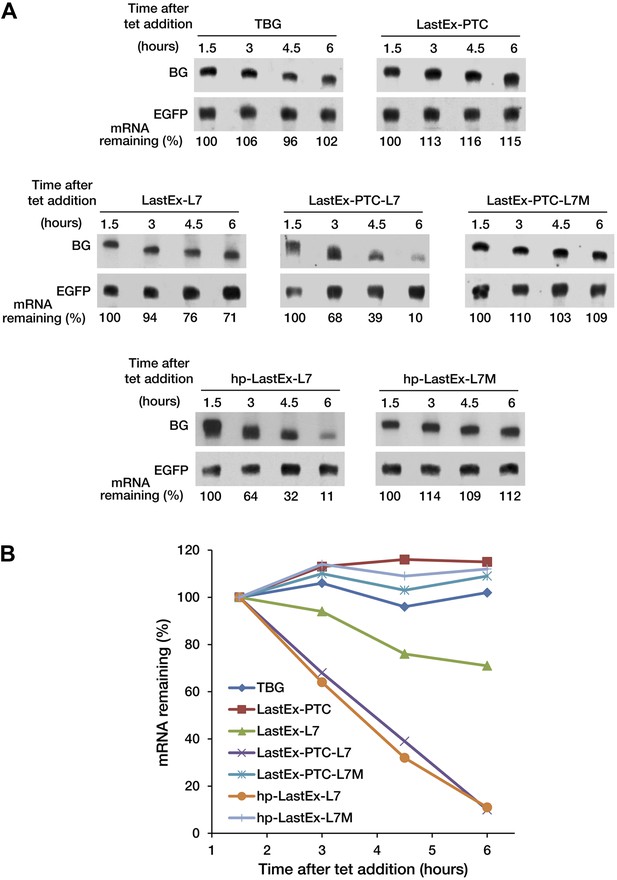
A PTC potentiates rapid miRNA-mediated decay of nonsense mRNAs.
(A) Decay of β-globin (BG) mRNAs. Cytoplasmic RNA was collected at the indicated times after transcriptional arrest by adding tetracycline (tet) and analyzed by Northern blotting. To more accurately quantify the half-life of BG mRNA, RNA samples were collected at later times than when monitoring mRNA deadenylation in Figure 1. (B) Graphs of the concentration of each BG mRNA in A as a function of time. All values were normalized to EGFP mRNA, a co-transfected constitutively transcribed internal standard.
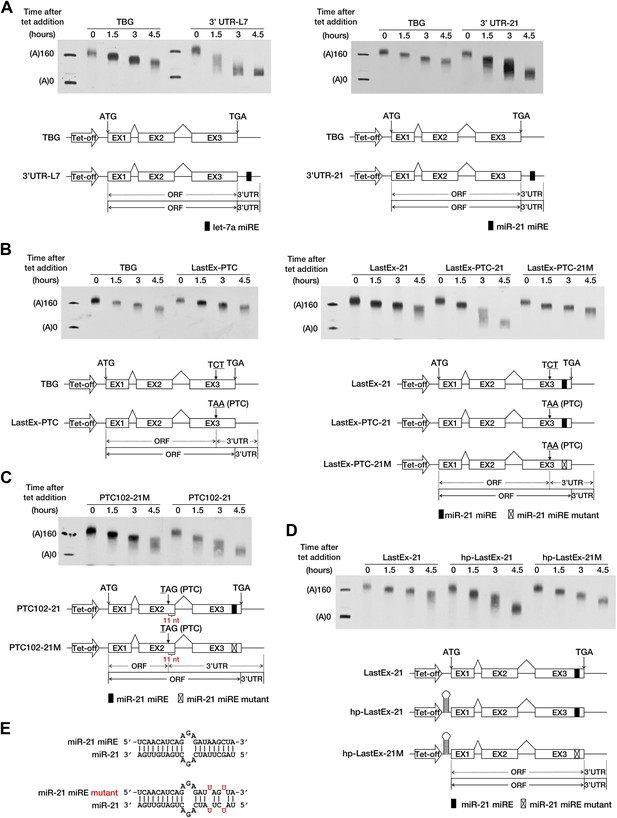
A PTC potentiates rapid miR-21-mediated deadenylation of nonsense mRNAs.
(A) Left panel: the influence of let-7a on the deadenylation rate of BG mRNA harboring a let-7a miRE in the 3′ UTR. Right panel: the influence of miR-21 on the deadenylation rate of BG mRNA harboring a miR-21 miRE in the 3′ UTR. (B) Left panel: deadenylation of BG mRNAs with or without a PTC in the last exon. TBG contains an intact BG ORF. A PTC was introduced into TBG at codon 116 within the last exon to generate LastEx-PTC. Neither construct harbors an miRE in its ORFs. Right panel: the influence of miR-21 on the deadenylation rate of BG mRNAs harboring one miR-21 miRE in its ORF with or without an upstream PTC mutation. LastEx-21 contains one miR-21 miRE in the last exon of the BG ORF. LastEx-PTC-21 has a PTC at the same position as LastEx-PTC, which is 22 nt upstream of the miR-21 miRE. Two nucleotides within the miR-21 miRE seed of LastEx-PTC-21 were mutated to create a synonymous codon in LastEx-PTC-21M. The positions of the ORF start site and the original stop codon are indicated by arrows. The borders of the original ORF/3′ UTR and redefined ORF/3′ UTR upon PTC mutation are indicated by solid or dashed lines below the constructs. (C) miRNA-induced accelerated deadenylation of nonsense BG mRNA that does not conform to the ‘50 nt boundary rule’ of EJC-NMD. PTC102-21 contains a PTC 11 nt upstream of the last exon–exon junction and a miR-21 miRE in the last exon of the ORF. PTC102-21M is identical to PTC102-21 except for a mutated miR-21 miRE. (D) The influence of miR-21 on the deadenylation rate of BG mRNA harboring a miR-21 miRE in its ORF in the absence of translation. hp-LastEx-21 contains a 40-nt inverted repeat in its 5′ UTR to block translation initiation. hp-LastEx-21M is identical to hp-LastEx-21 except for a mutated miR-21 miRE. (E) Duplexes expected for the miR-21 miRE or its mutant counterpart base-paired with miR-21.
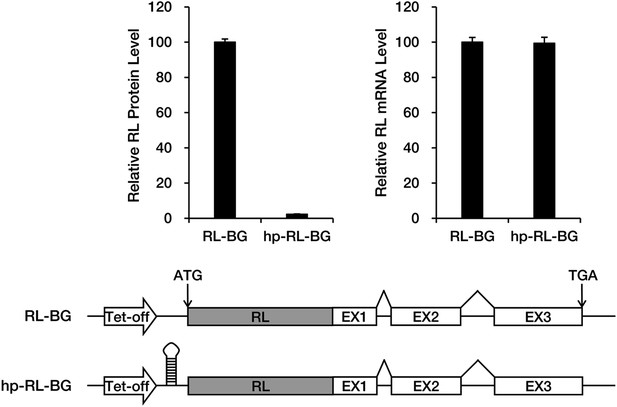
A large stem-loop structure in the 5′ UTR blocks translation of BG mRNA.
A Renilla luciferase (RL) ORF was fused to the 5′ side of the BG ORF so as to facilitate precise quantification of protein production. Levels of the RL-BG protein and mRNA were determined by assaying luciferase activity and by qRT-PCR, respectively. Amounts of RL protein and mRNA were normalized to a co-transfected firefly luciferase control.
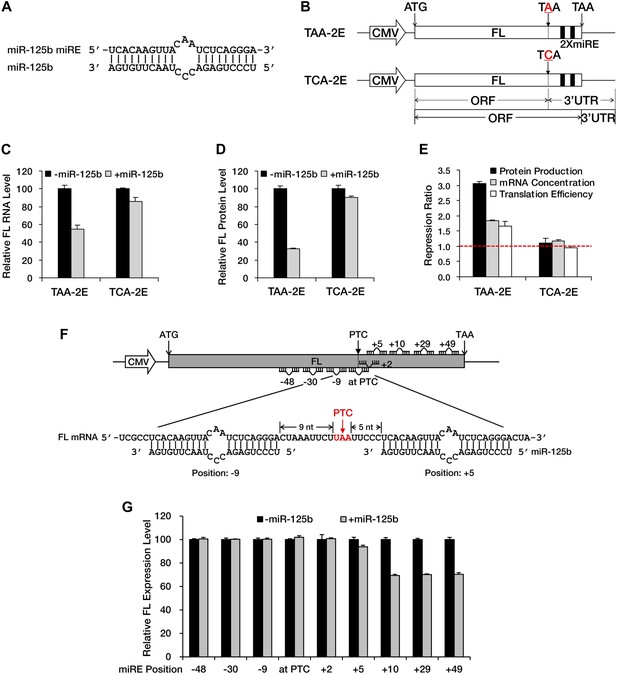
A PTC potentiates miRNA-mediated translational repression of nonsense mRNAs.
(A) Duplex expected for the miR-125b miRE base-paired with miR-125b. (B) The reporter constructs used in C to E. TAA-2E contains two miR-125b miREs 69 nt downstream of the original stop codon (which now serves as a PTC) of the firefly luciferase (FL) gene. This PTC (TAA) was mutated to codon TCA to generate TCA-2E. The borders of the ORF/3′ UTR and redefined ORF/3′ UTR upon PTC mutation are indicated by solid or dashed lines below the constructs. (C) mRNA quantification for TAA-2E and TCA-2E in the presence and absence of miR-125b, as determined by qRT-PCR. The error bars represent the standard deviation of multiple measurements. (D) Protein quantification by analyzing luciferase activity for TAA-2E and TCA-2E in the presence and absence of miR-125b. (E) Contribution of translational repression by miRNA-mediated surveillance. The repression ratios for TAA-2E and TCA-2E were calculated from normalized levels of firefly luciferase protein (black bars) and mRNA (gray bars) in the absence versus the presence of miR-125b. By dividing the repression ratio for protein production and that for mRNA concentration, the repression ratio for translation efficiency (protein yield per mRNA molecule, white bars) was determined. A repression ratio for translation efficiency that is >1 indicates that part of the total repression observed at the protein level is attributable to inhibition of translation. (F) Schematic illustration of the reporter designs used in G. The same 22-nt miR-125b miRE as in A was introduced in-frame into different positions before or after a PTC of a modified firefly luciferase reporter gene to obtain a series of miRE-containing plasmids. A graph that illustrates the different methods for calculating the distance between an upstream or a downstream miRE and the PTC is shown below the construct. (G) Boundary rule for miRNA-mediated surveillance. Each firefly luciferase construct that contains one miR-125b miRE at a different position was co-transfected with a Renilla luciferase reporter into HEK293 cells in the presence or absence of miR-125b. The relative FL expression level was calculated from the normalized levels of firefly luciferase in the absence versus the presence of miR-125b.
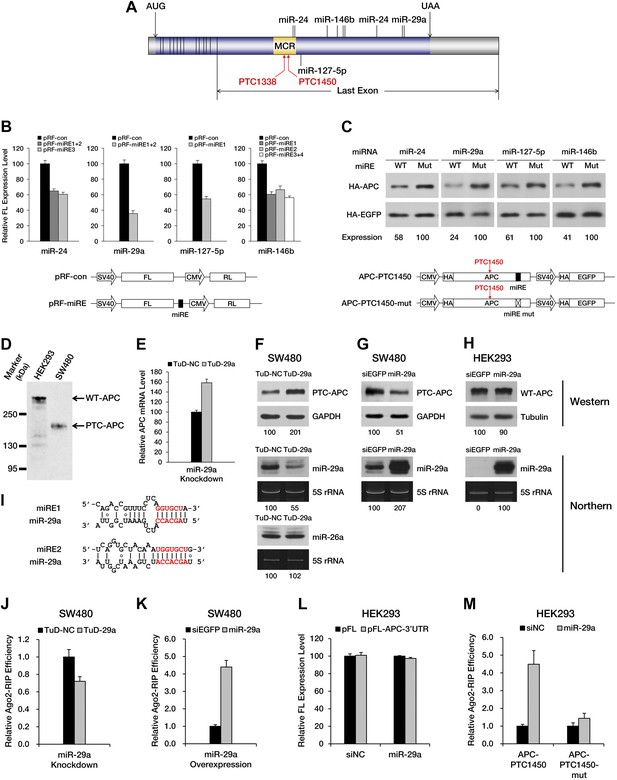
PTC-containing APC mRNAs are natural substrates repressed by miRNA-mediated surveillance.
(A) Schematic representation of APC mRNA. The ORF is shown in blue. The 5′ UTR and 3′ UTR are shown in gray. Exon–exon boundaries are indicated by vertical lines in the mRNA. The positions of representative miREs and PTC sites are indicated above or below the mRNA by solid lines and arrows, respectively. MCR refers to the mutation cluster region. (B) Validation of miRE function by luciferase assays. A vector expressing both a firefly luciferase (FL) transcript harboring one potential miRE in its 3′ UTR and a control Renilla luciferase (RL) transcript was co-transfected with cognate miRNA mimics into HEK293 cells. The relative FL expression level represents the firefly/Renilla luciferase ratio for pRF-miRE relative to the no miRE control pRF-con. (C) Validation of miRE function in an APC minigene. A vector expressing both an HA-tagged truncated APC (APC-PTC1450) and a control HA-tagged EGFP was co-transfected with cognate miRNA mimics into HEK293 cells. The mutant counterpart of the APC minigene (APC-PTC1450-mut) contains two altered nucleotides that abolish miRE:miRNA complementarity without changing the identity of the encoded amino acid. (D) Western blot analysis of endogenous truncated APC in SW480 cells and full-length APC in HEK293 cells by using an anti-APC antibody. (E) Change in PTC-APC mRNA levels in SW480 cells upon miR-29a knockdown. Cytoplasmic RNA was extracted from the SW480 cell lines used in F, and the levels of APC and GAPDH mRNA were determined by qRT-PCR. The relative APC mRNA level was calculated by normalizing to GAPDH mRNA. (F) Upregulation of endogenous truncated APC upon miR-29a knockdown. SW480 cells were transduced with lentiviruses encoding a miR-29a decoy (TuD-29a) or a control decoy (TuD-NC). Endogenous truncated APC was probed with an anti-APC antibody. GAPDH served as a loading control. Changes in the levels of endogenous miR-29a and an untargeted control (miR-26a) were determined by Northern blotting. 5S rRNA served as a loading control. (G) Downregulation of endogenous truncated APC upon miR-29a overexpression. SW480 cells were transduced with lentiviruses encoding miR-29a or a control small RNA (siEGFP). Western and Northern assays were performed as in F. (H) Invariant concentration of endogenous wild-type APC upon miR-29a overexpression. HEK293 cells were transduced with lentiviruses encoding miR-29a or a control small RNA (siEGFP). Western and Northern assays were performed as in F except that Tubulin served as the loading control in the Western assay. (I) Duplexes expected for the miR-29a miREs base-paired with miR-29a. (J) Ribonucleoprotein immunoprecipitation (RIP) analysis of PTC-APC mRNA associated with Ago2 in SW480 cells upon miR-29a knockdown. SW480 cells used in F were transduced with a low amount of lentiviruses (MOI <0.3) expressing FLAG-tagged Ago2. Anti-FLAG RIP followed by qRT-PCR was performed to compare the binding of endogenous PTC-APC mRNAs to Ago2. The amount of Ago2-associated PTC-APC mRNA was normalized to MYC, an endogenous target of let-7c. The relative Ago2-RIP efficiency was calculated from the normalized amount of PTC-APC mRNA in the presence of a miR-29a decoy (TuD-29a) versus a control decoy (TuD-NC). (K) RIP analysis of PTC-APC mRNA associated with Ago2 in SW480 cells upon miR-29a overexpression. SW480 cells used in G were transduced with a low amount of lentiviruses (MOI <0.3) expressing FLAG-tagged Ago2. RIP assays were performed as in J. The relative Ago2-RIP efficiency was calculated from the normalized amount of Ago2-associated PTC-APC mRNA in the presence of miR-29a versus a control small RNA (siEGFP). (L) The 3′ UTR of APC mRNA contains no miR-29a miRE. The sequence of a full length APC 3′ UTR was cloned to the 3′ UTR of a firefly luciferase reporter. This reporter plasmid (pFL-APC-3′UTR) or a control plasmid (pFL) was co-transfected with a miR-29a mimic or a control small RNA (siNC) into HEK293 cells. The relative FL expression level was calculated from the normalized levels of firefly luciferase activity for pFL-APC-3′UTR versus pFL in the presence of the miR-29a mimic or siNC. (M) RIP analysis of ectopically expressed PTC-APC mRNA associated with Ago2 in HEK293 cells. A PTC-containing APC minigene plasmid with wild-type (APC-PTC1450) or mutant miR-29a miREs (APC-PTC1450-mut) was co-transfected with the miR-29a mimic or a control small RNA (siNC) into HEK293 cells that stably expressed FLAG-tagged Ago2. Anti-FLAG RIP followed by qRT-PCR was performed to compare the binding of the mRNAs to Ago2. The amount of Ago2-associated PTC-APC mRNA or its miRE mutant counterpart was normalized to HOXD10, an endogenous target of miR-10a. The relative Ago2-RIP efficiency was calculated from the normalized levels of Ago2-associated PTC-APC mRNA bearing wild-type (APC-PTC1450) or mutant (APC-PTC1450-mut) miR-29a miREs in the presence of the miR-29a mimic versus siNC.
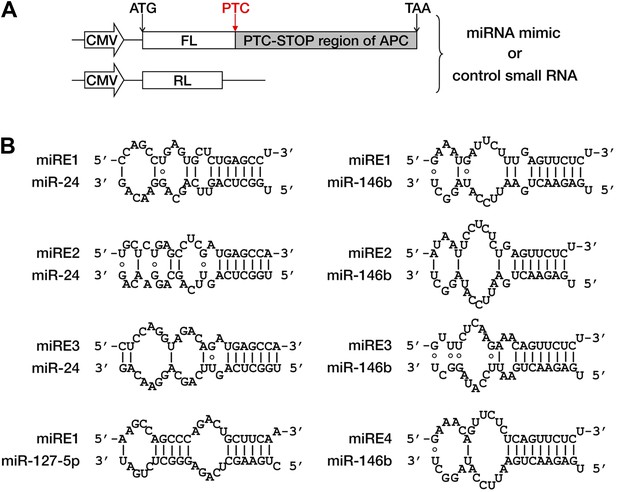
Duplexes expected for functional miRNAs base-paired with cognate miREs.
(A) Reporter constructs and strategy for miRE screening of APC mRNA. The cDNA region between codon 1450 and the stop codon of APC mRNA was fused to the 3′ end of the firefly luciferase (FL) ORF such that the original stop codon of the luciferase gene functioned as a PTC. The PTC-STOP region of APC mRNA was examined for predicted miREs. To test each putative miRE, a cognate miRNA mimic and a control small RNA (siEGFP) were co-transfected with the reporter into HEK293 cells. As an internal control, a Renilla luciferase (RL) reporter was simultaneously transfected. miRNAs that caused strong repression were selected for further validation. (B) Duplexes expected for miR-24, miR-127-5p, and miR-146b base-paired with their cognate miREs.
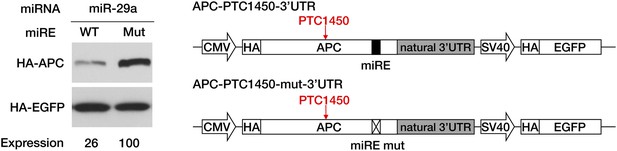
Repressive activity of miR-29a miREs within the PTC-STOP region of APC minigenes bearing the natural APC 3′ UTR.
The constructs were the same as in Figure 3C except that they contained the full length 3′ UTR of APC mRNA. Western assays were performed as in Figure 3C.
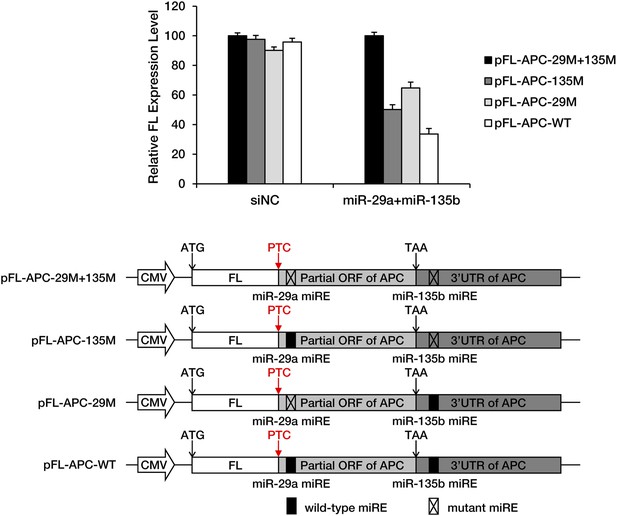
Full functionality of miREs in the PTC-STOP region in the presence of 3′ UTR-dependent repression by miRNAs.
The PTC-STOP region and the entire 3′ UTR of APC mRNA were fused to the 3′ end of a firefly luciferase (FL) ORF so that the stop codon of luciferase served as a PTC. miREs for miR-29a (ORF miRE) and miR-135b (3′ UTR miRE) were mutated individually or simultaneously. HEK293 cells were co-transfected with each reporter together with equal amounts of miR-29a and miR-135b or a control small RNA (siNC). Relative FL expression represents the ratio of firefly/Renilla luciferase for each reporter relative to pFL-APC-29M+135M.
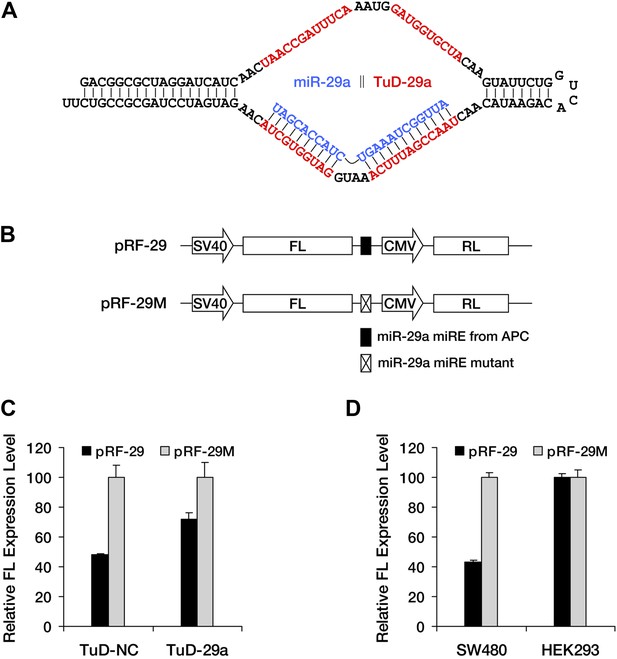
Successful knockdown of endogenous miR-29a by TuD-29a.
(A) Schematic representation of the structure of the TuD decoy RNA for miR-29a. (B) Reporter constructs used in C and D. pRF-29 contains the miR-29a miRE originating from APC, whereas pRF-29M contains a mutant form of the miR-29a miRE. Both reporters express Renilla luciferase (RL) from a second promoter in the same construct as a control. (C) Successful knockdown of endogenous miR-29a in SW480 cells by TuD-29a. The relative expression level of pRF-29 was elevated in TuD-29a-expressing cells compared to TuD-NC-expressing control cells. The relative FL expression level was calculated from the normalized levels of firefly luciferase activity for pRF-29 versus pRF-29M. (D) Tissue-specific effect of miRNA-mediated surveillance. pRF-29 is efficiently repressed by endogenous miR-29a in colon-derived SW480 cells but not in kidney-derived HEK293 cells. The relative FL expression level was calculated as in C.

Confirmation of interaction between miR-29a and its miREs in APC mRNA.
A mutant miR-29a mimic that restored seed complementarity with the mutant miR-29a miREs was co-transfected with APC-PTC1450 minigene plasmids bearing wild-type or mutant miR-29a miREs. Western assays were performed as in Figure 3C. The mutated nucleotides are underlined in blue.
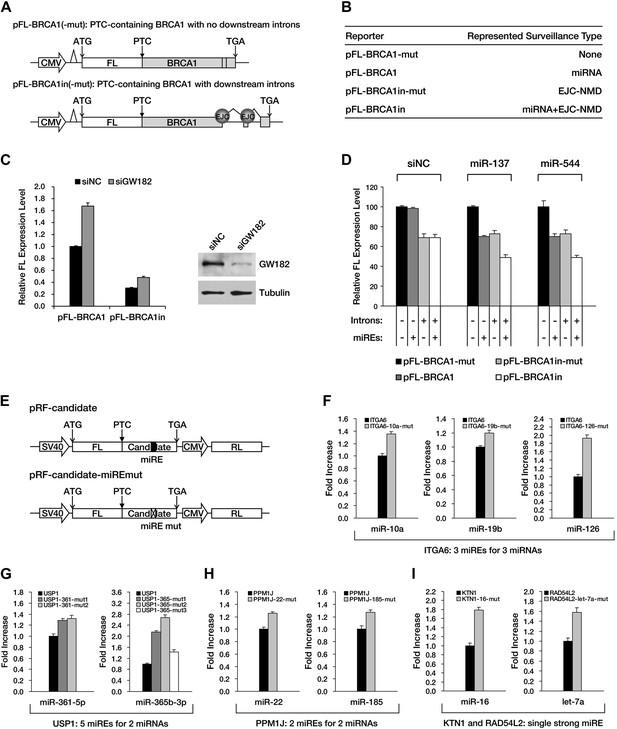
The repressive effects of miRNA-mediated surveillance and EJC-NMD are additive.
(A) The reporter constructs used in C and D. pFL-BRCA1 and pFL-BRCA1-mut have no introns, whereas pFL-BRCA1in and pFL-BRCA1in-mut each harbor two introns downstream of PTC. The miREs for miR-137 and miR-544 were mutated to synonymous codons in pFL-BRCA1-mut and pFL-BRCA1in-mut. (B) Identification of the surveillance mechanism pertinent to each reporter. (C) Left panel: miRNA-mediated surveillance and EJC-NMD are not mutually exclusive. Knockdown of RISC core component GW182 alleviated the repression of BRCA1 reporters, regardless of the presence of the downstream introns. The relative FL expression level represents the firefly/Renilla luciferase ratio in the presence of a control siRNA (siNC) versus an siRNA targeting GW182 (siGW182). Right panel: expression level of GW182 protein in HeLa-tTA cells treated with siNC or siGW182. Tubulin served as a loading control. (D) Additive effects of miRNA-mediated surveillance and EJC-NMD. Each reporter in A was co-transfected with miR-137, miR-544, or a control small RNA (siNC), together with a Renilla luciferase reporter into HEK293 cells. The relative FL expression level represents the firefly/Renilla luciferase ratio of each EJC-NMD- and/or miRNA-responsive reporter relative to pFL-BRCA1-mut. (E) Reporter constructs for miRE function validation of candidates identified from HCT-116 exome and RNA sequencing data. The PTC-STOP region of each candidate was fused to a firefly luciferase (FL) ORF in the same manner as for APC and BRCA1. A control Renilla luciferase (RL) reporter was expressed from a second promoter in the same construct (pRF-candidate). Each potential miRE identified from the screening was mutated to obtain a series of miRE mutant reporters (pRF-candidate-miREmut). (F–I) Experimental verification of functional miREs in the PTC-STOP region of selected candidates. The wild-type or miRE mutant version of each candidate reporter was co-transfected into HEK293 cells with a cognate miRNA mimic. The activity of each miRE (fold increase) was calculated from the normalized levels of firefly luciferase activity for pRF-candidate versus pRF-candidate-miREmut in the presence of the cognate miRNA mimic.
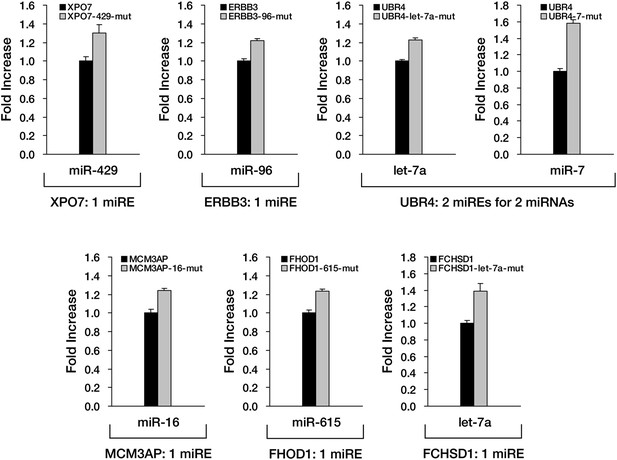
Experimental verification of functional miREs in the PTC-STOP region of additional nonsense mutant mRNAs.
Six additional candidates identified from HCT-116 cells were subjected to experimental verification of predicted miREs in their PTC-STOP regions. The wild-type or miRE mutant version of the reporter for each candidate was co-transfected into HEK293 cells with a cognate miRNA mimic. The activity of each miRE was calculated as in Figure 4F–I.
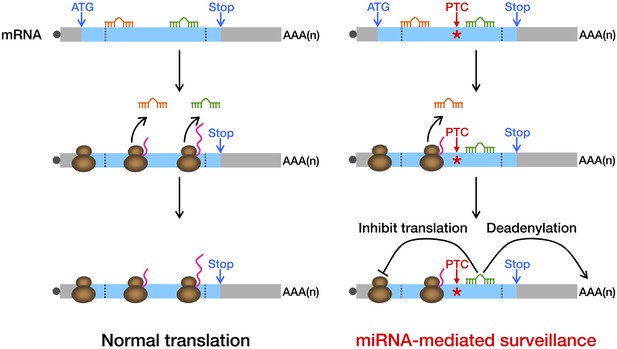
Model of miRNA-mediated surveillance system.
The coding region of an mRNA may contain multiple potential miREs. Usually, miRNAs cannot stably bind to their cognate miREs that are embedded within the ORF of a normal transcript under active translation. However, upon nonsense mutation, the translating ribosome stalls at the PTC so that miREs downstream of the PTC are unmasked, triggering miRNA-mediated deadenylation and translational repression.
Additional files
-
Supplementary file 1
Screening results of functional miREs in the PTC-STOP region of APC mRNA. Reporter constructs and the strategy for miRE screening were described in Figure 3—figure supplement 1A. In total, 45 miRNAs based on their general abundance in human tissues and the predicted thermal stability of the duplex they may form with an APC miRE were chosen for the screening. The repression ratio for each miRNA was calculated from normalized levels of firefly luciferase activity in the presence of a control small RNA (siEGFP) versus a miRNA mimic.
- https://doi.org/10.7554/eLife.03032.017
-
Supplementary file 2
Transcriptome-wide identification of nonsense mRNAs caused by point mutation in HCT-116 cells. Exome sequencing was performed to identify all PTC-containing mutants in the genome of HCT-116 cells. Genes with the PTC/WT allele reads ratio between 0.7 and 1.3 were considered to be heterozygous. Transcriptome sequencing data were then analyzed to evaluate the effect of these heterozygous PTCs on mRNA stability. Sequencing reads that specifically mapped to the PTC and wild-type allele were counted and used to calculate the relative concentration of the respective transcripts. In total, 47 heterozygous PTC-containing mRNAs predicted from the genomic sequence were actively expressed in HCT-116 cells, as shown in the list. Candidates were subdivided into EJC-NMD-sensitive and/or miRNA-mediated surveillance-sensitive groups according to the position of the PTC. miREs in the PTC-STOP region of each candidate were predicted based on the 2–7 seed match rule and the top 150 expressed miRNAs in HCT-116 cells. Candidates highlighted in yellow were selected for experimental validation. Candidates that contain at least one experimentally verified miRE are shown in red font.
- https://doi.org/10.7554/eLife.03032.018
-
Supplementary file 3
Transcriptome-wide identification of nonsense mRNAs caused by intron retention in HEK293 and HeLa cells. The isoform with the highest transcript abundance in each gene family was chosen as the constitutive isoform. All intron-retention (IR) isoforms with an intron retention level >0.05 were then found by using MATS. Only PTC-causing IR candidates with Ago-CLIP hits in their PTC-STOP regions were listed. miREs were predicted in the PTC-STOP region using the 2–7 seed match rule. Candidates were subdivided into EJC-NMD-sensitive and/or miRNA-mediated surveillance-sensitive groups.
- https://doi.org/10.7554/eLife.03032.019





