Cryo-EM structure of the Plasmodium falciparum 80S ribosome bound to the anti-protozoan drug emetine
Figures
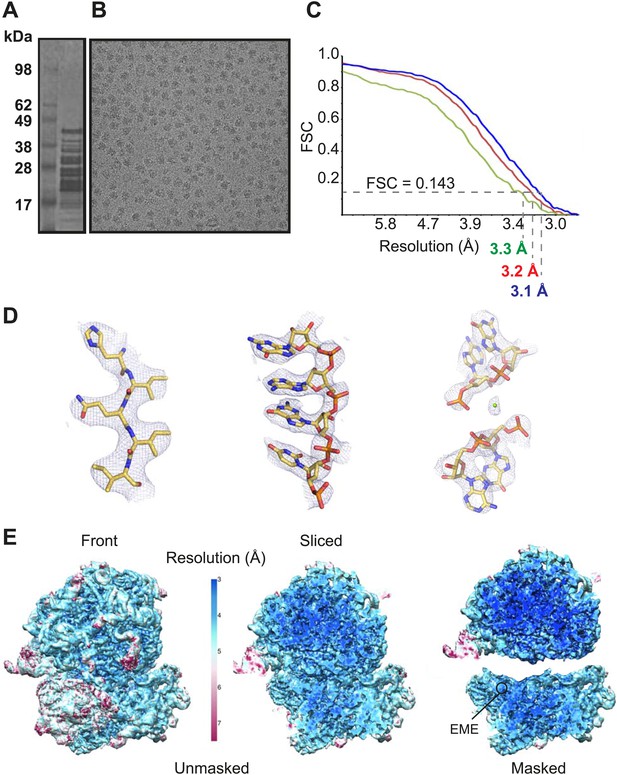
Cryo-EM data and processing.
(A) Sucrose gradient purification of Pf80S ribosomes. (B) Representative electron micrograph showing Pf80S particles. (C) Fourier Shell Correlation (FSC) curves indicating the overall resolutions of unmasked (red), Pf40S masked (green) and Pf60S masked (blue) reconstructions of the Pf80S–emetine complex. (D) Representative density with built models of a β-strand with well-resolved side chains (left), an RNA segment with separated bases (middle), and a magnesium ion (green sphere) that is coordinated by RNA backbone phosphates. (E) Density maps colored according to local resolution for the unmasked Pf80S (left) and masked Pf40S and Pf60S subunits (right).
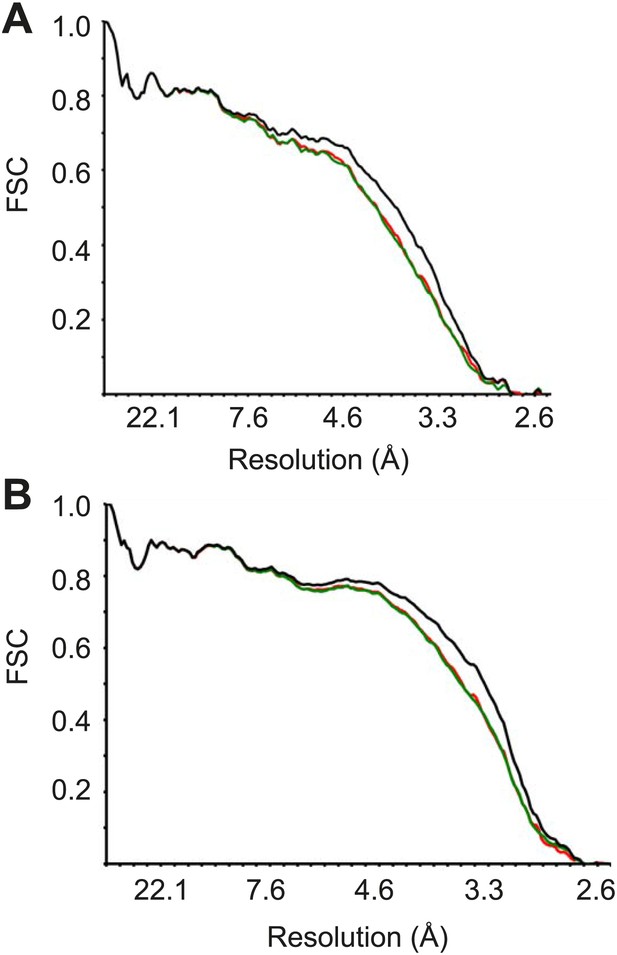
FSC curves between the final refined atomic model and the reconstructions from all particles (black); between the model refined in the reconstruction from only half of the particles and the reconstruction from that same half (FSCwork, red); and between that same model and the reconstruction from the other half of the particles (FSCtest, green), for Pf40S (A) and Pf60S (B).
https://doi.org/10.7554/eLife.03080.004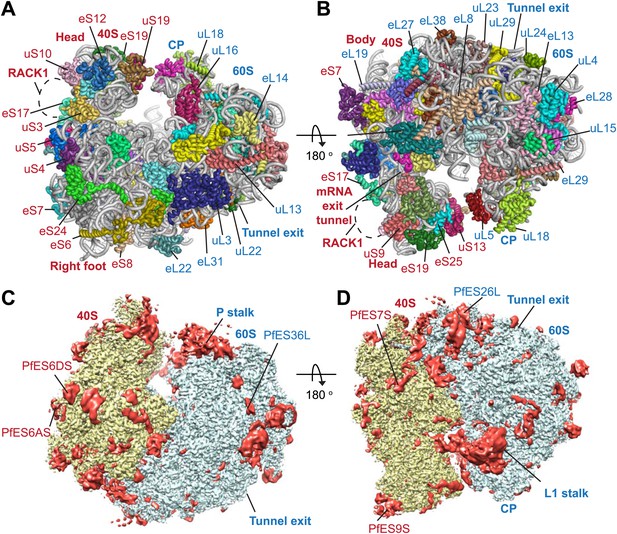
Structure of the Pf80S ribosome.
Overview of Pf80S atomic model showing views facing (A) tRNA entry side and (B) tRNA exit side. rRNAs are shown in gray, proteins numbered according to Ban et al. (2014). (C and D) Pf40S and Pf60S subunits are colored in yellow and blue respectively. Flexible regions are shown in red and at a resolution of 6 Å. Pf-specific expansion segments (ESs) relative to human ribosomes are labeled. Their numbering is as described for the human cytoplasmic ribosome (Anger et al., 2013).
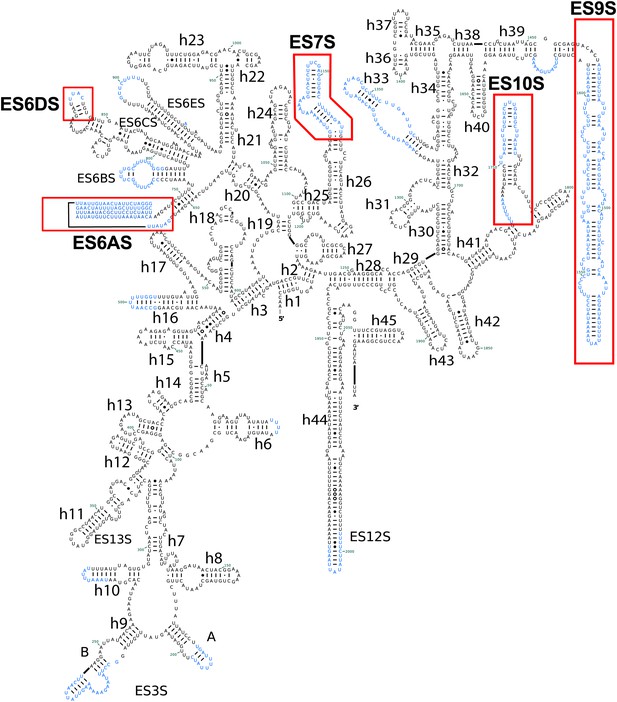
Secondary structure of Pf18S rRNAs.
Pf-specific ESs are highlighted in a labeled red box. Regions not built in the atomic model are colored in blue text. The secondary structure was modified from the CRW site (Cannone et al., 2002).
Secondary structure of the 5′ half of Pf 28S rRNA.
Pf-specific ESs are highlighted in a labeled red box. Regions not built in the atomic model are colored in blue text. The secondary structure was modified from the CRW site (Cannone et al., 2002).
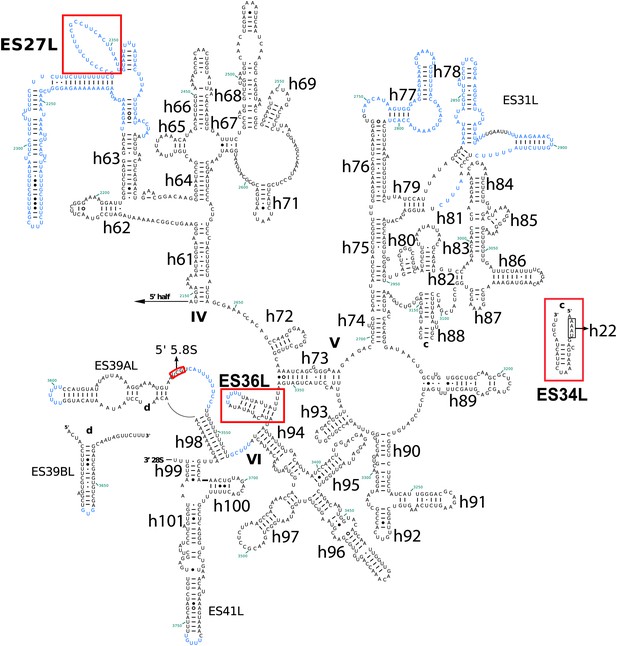
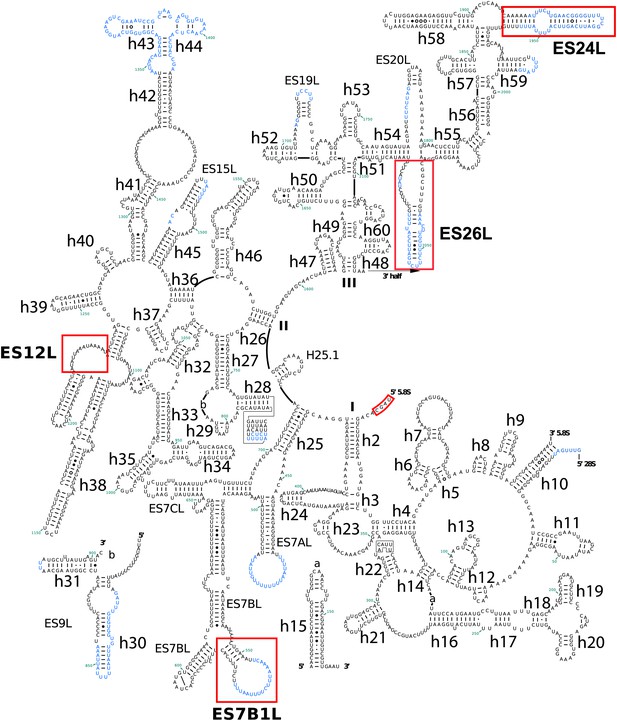
Secondary structure of the 3′ half of Pf28S rRNA.
Pf-specific ESs are highlighted in a labeled red box. Regions not built in the atomic model are colored in blue text. The secondary structure was modified from the CRW site (Cannone et al., 2002).
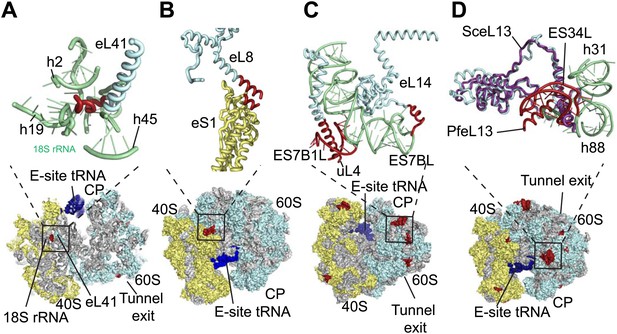
Details of Pf-specific protein extensions and rRNA ESs near the (A and B) subunit interface (C) P stalk and (D) the L1 stalk.
Pf-specific elements are shown in red.
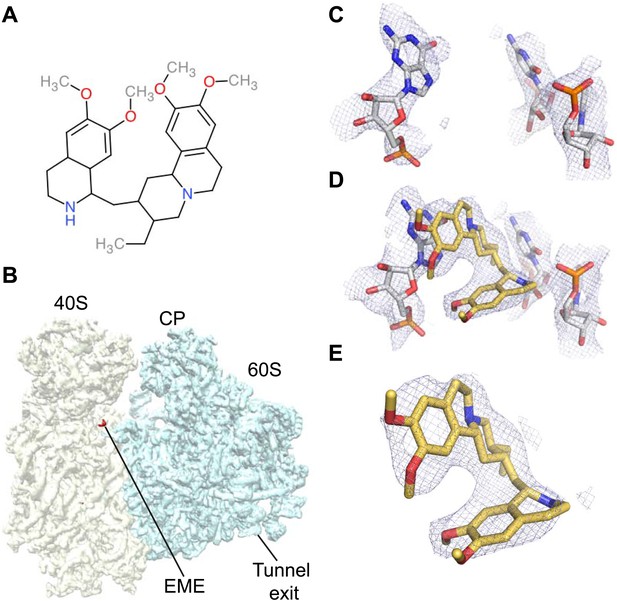
Emetine binds to the E-site of the Pf40S subunit.
(A) 2D chemical structure of emetine. (B) A 4.5 Å filtered difference map (red density) at 5 standard deviation overlaid with the Pf80S map filtered at 6 Å (blue and yellow for Pf60S and Pf40S respectively), showing the emetine density at the E-site of the Pf40S. The emetine binding site in (C) empty and (D) emetine-bound structures, with (E) density for emetine alone at 3.2 Å.
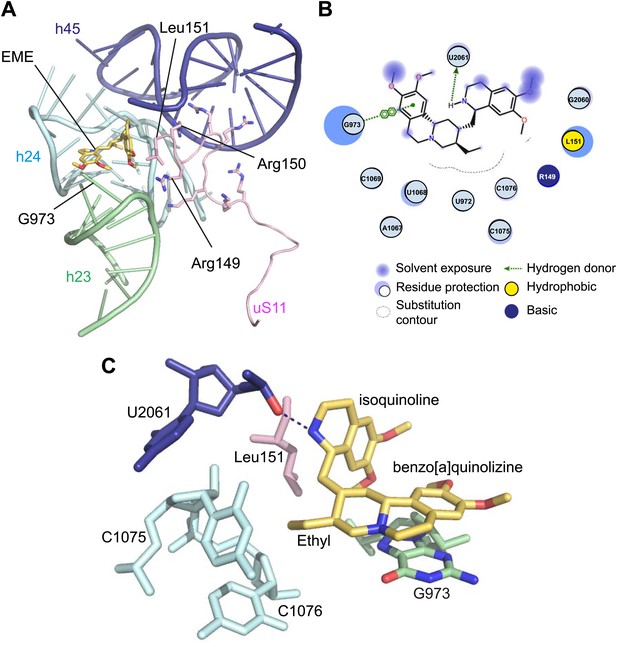
Molecular details of the emetine–ribosome interaction.
(A) Overview of emetine at the binding interface formed by the three conserved rRNA helices and uS11. h23 is in green, h24 in cyan, h45 in blue, uS11 in pink, and emetine in yellow. (B) 2D representation showing the interaction of emetine with binding residues. Substitution contour represents potential space for chemical modification of emetine. (C) Residues in physical contact with emetine. Hydrogen bond is indicated as dashes.
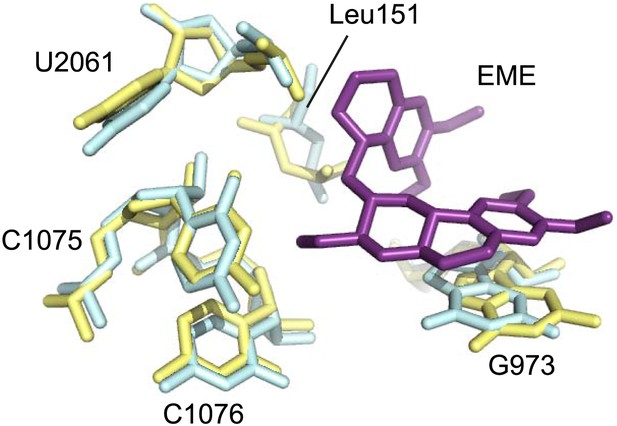
Comparison of the emetine binding residues between Pf80S and human ribosomes.
Human and Pf-specific elements are colored in yellow and cyan respectively, with Pf numbering. Emetine is in purple.
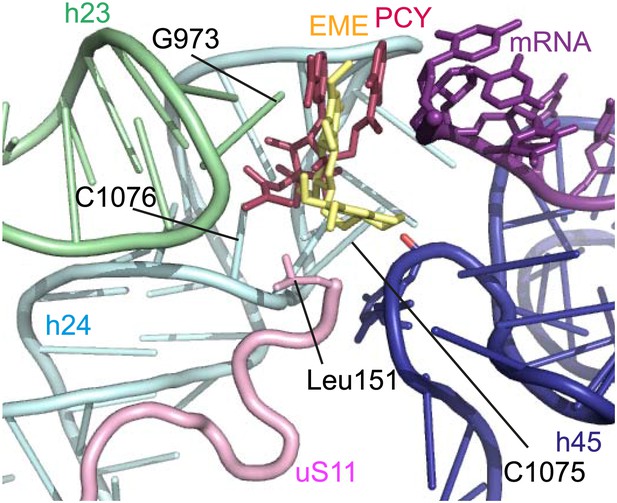
Comparison with pactamycin.
Superposition of emetine and pactamycin at the Pf40S emetine binding pocket. Emetine and pactamycin are shown in yellow and red respectively.
Tables
Refinement and model statistics
| Pf80S–emetine | |
|---|---|
| Data collection | |
| Particles | 105,247 |
| Pixel size (Å) | 1.34 |
| Defocus range (μm) | 0.8–3.8 |
| Voltage (kV) | 300 |
| Electron dose (e− Å−2) | 20 |
| Pf60S | Pf40S | |
|---|---|---|
| Model composition | ||
| Non-hydrogen atoms | 124,509 | 68,858 |
| Protein residues | 6,244 | 4,106 |
| RNA bases | 3,460 | 1,682 |
| Ligands (Zn2+/Mg2+/emetine) | 5/163/0 | 1/67/1 |
| Refinement | ||
| Resolution used for refinement (Å) | 3.1 | 3.3 |
| Map sharpening B-factor (Å2) | −60.3 | −79.9 |
| Average B factor (Å2) | 113.1 | 143.2 |
| Rfactor* | 0.2294 | 0.257 |
| Fourier Shell Correlation† | 0.86 | 0.854 |
| Rms deviations | ||
| Bonds (Å) | 0.006 | 0.007 |
| Angles (°) | 1.20 | 1.29 |
| Validation (proteins) | ||
| Molprobity score | 2.45 (96th percentile) | 2.73 (95th percentile) |
| Clashscore, all atoms | 3.65 (100th percentile) | 4.23 (100th percentile) |
| Good rotamers (%) | 90.0 | 86.0 |
| Ramachandran plot | ||
| Favored (%) | 90.4 | 85.4 |
| Outliers (%) | 2.4 | 4.2 |
| Validation (RNA) | ||
| Correct sugar puckers (%) | 97.3 | 97.5 |
| Good backbone conformations (%) | 71.1 | 70.0 |
-
*
Rfactor = Σ||Fobs| − ||Fcalc|/Σ|Fobs|.
-
†
FSCoverall = Σ(Nshell FSCshell)/Σ(Nshell), where FSCshell is the FSC in a given shell, Nshell is the number of ‘structure factors’ in the shell. FSCshell = Σ(Fmodel FEM)/(√(Σ(|F|2model)) √(Σ(F2EM)).
Ribosomal proteins of the Pf40S subunit
| Protein names | Uniprot ID | PlasmoDB ID | Chain ID | Built residues | Extensions compared to human | Total number of residues |
|---|---|---|---|---|---|---|
| eS1 | RS3A_PLAF7 | PF3D7_0322900 | B | 24–233 | 245–262 | 262 |
| uS2 | RSSA_PLAF7 | PF3D7_1026800 | C | 10–204 | – | 263 |
| uS3 | Q8IKH8_PLAF7 | PF3D7_1465900 | D | 4–39; 65–78; 97–193; 207–216 | – | 221 |
| uS4 | Q8I3R0_PLAF7 | PF3D7_0520000 | E | 2–186 | – | 189 |
| eS4 | Q8IIU8_PLAF7 | PF3D7_1105400 | F | 2–258 | – | 261 |
| uS5 | Q8IL02_PLAF7 | PF3D7_1447000 | G | 39–262 | – | 272 |
| eS6 | Q8IDR9_PLAF7 | PF3D7_1342000 | H | 1–160; 170–213 | 249–306 | 306 |
| uS7 | Q8IBN5_PLAF7 | PF3D7_0721600 | I | 7–118; 128–195 | – | 195 |
| eS7 | Q8IET7_PLAF7 | PF3D7_1302800 | J | 3–190 | – | 194 |
| uS8 | O77395_PLAF7 | PF3D7_0316800 | K | 2–130 | – | 130 |
| eS8 | Q8IM10_PLAF7 | PF3D7_1408600 | L | 5–120; 161–213; 216–218 | 154–163 | 218 |
| uS9 | Q8IAX5_PLAF7 | PF3D7_0813900 | M | 6–143 | – | 144 |
| uS10 | Q8IK02_PLAF7 | PF3D7_1003500 | N | 21–118 | – | 118 |
| eS10 | Q8IBQ5_PLAF7 | PF3D7_0719700 | O | 11–89 | – | 137 |
| uS11 | Q8I3U6_PLAF7 | PF3D7_0516200 | P | 25–151 | – | 151 |
| uS12 | O97248_PLAF7 | PF3D7_0306900 | Q | 2–145 | – | 145 |
| eS12 | RS12_PLAF7 | PF3D7_0307100 | R | 22–78; 85–100; 111–135 | 10–16 | 141 |
| uS13 | Q8IIA2_PLAF7 | PF3D7_1126200 | S | 12–139 | – | 156 |
| uS14 | C0H4K8_PLAF7 | PF3D7_0705700 | T | 7–54 | – | 54 |
| uS15 | Q8IDB0_PLAF7 | PF3D7_1358800 | U | 3–151 | – | 151 |
| uS17 | O77381_PLAF7 | PF3D7_0317600 | V | 6–25; 36–161 | – | 161 |
| eS17 | Q8I502_PLAF7 | PF3D7_1242700 | W | 3–83; 97–110 | – | 137 |
| uS19 | C0H5C2_PLAF7 | PF3D7_1317800 | X | 21–95; 103–123 | – | 145 |
| eS19 | Q8IFP2_PLAF7 | PF3D7_0422400 | Y | 15–168 | 1–19 | 170 |
| eS21 | Q8IHS5_PLAF7 | PF3D7_1144000 | Z | 11–82 | – | 82 |
| eS24 | Q8I3R6_PLAF7 | PF3D7_0519400 | 1 | 3–122 | – | 133 |
| eS25 | Q8ILN8_PLAF7 | PF3D7_1421200 | 2 | 35–42; 58–84; 97–102 | – | 105 |
| eS26 | O96258_PLAF7 | PF3D7_0217800 | 3 | 2–96 | – | 107 |
| eS27 | Q8IEN2_PLAF7 | PF3D7_1308300 | 4 | 7–82 | – | 82 |
| eS28 | Q8IKL9_PLAF7 | PF3D7_1461300 | 5 | 2–29; 37–66 | – | 67 |
| eS30 | RS30_PLAF7 | PF3D7_0219200 | 6 | 6–48 | – | 58 |
| eS31 | Q8IM64_PLAF7 | PF3D7_1402500 | – | Not built | – | 149 |
Ribosomal proteins of the Pf60S subunit
| Protein names | Uniprot ID | PlasmoDB ID | Chain ID | Built residues | Extensions compared to human | Total number of residues |
|---|---|---|---|---|---|---|
| uL2 | Q8I3T9_PLAF7 | PF3D7_0516900 | D | 2–248 | – | 260 |
| uL3 | Q8IJC6_PLAF7 | PF3D7_1027800 | E | 2–381 | – | 386 |
| uL4 | Q8I431_PLAF7 | PF3D7_0507100 | F | 6–395 | 373–411 | 411 |
| uL5 | Q8IBQ6_PLAF7 | PF3D7_0719600 | G | 8–51; 64–85; 92–106; 124–166 | – | 173 |
| uL6 | Q8IE85_PLAF7 | PF3D7_1323100 | H | 2–186 | – | 190 |
| eL6 | Q8IDV1_PLAF7 | PF3D7_1338200 | I | 9–151; 158–221 | 110–118; 139–143; 174–182 | 221 |
| eL8 | Q8ILL2_PLAF7 | PF3D7_1424400 | J | 40–46; 54–131; 147–283 | 11–24;279–283 | 283 |
| uL13 | Q8IJZ7_PLAF7 | PF3D7_1004000 | K | 1–201 | – | 202 |
| eL13 | Q8IAX6_PLAF7 | PF3D7_0814000 | L | 2–212 | 134–141; 168–174 | 215 |
| uL14 | Q8IE09_PLAF7 | PF3D7_1331800 | M | 8–139 | – | 139 |
| eL14 | Q8ILE8_PLAF7 | PF3D7_1431700 | N | 5–150 | 1–18 | 165 |
| uL15 | C6KT23_PLAF7 | PF3D7_0618300 | O | 2–148 | – | 148 |
| eL15 | C0H4A6_PLAF7 | PF3D7_0415900 | P | 2–205 | – | 205 |
| uL16 | Q8ILV2_PLAF7 | PF3D7_1414300 | Q | 2–101; 118–206 | – | 219 |
| uL18 | Q8ILL3_PLAF7 | PF3D7_1424100 | R | 5–126; 141–185; 189–250; 271–293 | – | 294 |
| eL18 | C0H5G3_PLAF7 | PF3D7_1341200 | U | 5–184 | – | 184 |
| eL19 | C6KSY6_PLAF7 | PF3D7_0614500 | T | 2–182 | – | 182 |
| eL20 | Q8IDS6_PLAF7 | PF3D7_1341200 | S | 2–187 | – | 184 |
| eL21 | Q8ILK3_PLAF7 | PF3D7_1426000 | V | 4–158 | – | 161 |
| uL22 | Q8IDI5_PLAF7 | PF3D7_1351400 | W | 4–154; 197–215 | – | 203 |
| eL22 | Q8IB51_PLAF7 | PF3D7_0821700 | X | 40–136 | 4–18; 34–38 | 139 |
| uL23 | Q8IE82_PLAF7 | PF3D7_1323400 | Y | 88–188 | 13–34; 57–67 | 190 |
| uL24 | O77364_PLAF7 | PF3D7_0312800 | Z | 2–122 | – | 126 |
| eL24 | Q8IEM3_PLAF7 | PF3D7_1309100 | 0 | 8–69 | – | 162 |
| eL27 | Q8IKM5_PLAF7 | PF3D7_1460700 | 1 | 2–126;132–146 | – | 146 |
| eL28 | Q8IHU0_PLAF7 | PF3D7_1142500 | 2 | 2–69; 77–82; 86–98; 103–119 | – | 127 |
| uL29 | Q8IIB4_PLAF7 | PF3D7_1124900 | 3 | 3–121 | – | 124 |
| eL29 | C6S3J6_PLAF7 | PF3D7_1460300 | 4 | 2–67 | – | 67 |
| uL30 | O97250_PLAF7 | PF3D7_0307200 | 5 | 35–257 | – | 257 |
| eL30 | Q8IJK8_PLAF7 | PF3D7_1019400 | 6 | 8–105 | – | 108 |
| eL31 | Q8I463_PLAF7 | PF3D7_0503800 | 7 | 15–88; 95–116 | – | 120 |
| eL32 | Q8I3B0_PLAF7 | PF3D7_0903900 | 8 | 2–126 | – | 131 |
| eL33 | Q8IHT9_PLAF7 | PF3D7_1142600 | 9 | 35–137 | 1–35 | 140 |
| eL34 | Q8IBY4_PLAF7 | PF3D7_0710600 | a | 2–107 | – | 150 |
| eL36 | Q8I713_PLAF7 | PF3D7_1109900 | b | 2–27; 38–106 | 5–10 | 112 |
| eL37 | C0H4L5_PLAF7 | PF3D7_0706400 | c | 2–90 | – | 92 |
| eL38 | Q8II62_PLAF7 | PF3D7_1130100 | d | 2–31; 36–77 | – | 87 |
| eL39 | C0H4H3_PLAF7 | PF3D7_0611700 | e | 2–30; 38–51 | – | 51 |
| eL40 | Q8ID50_PLAF7 | PF3D7_1365900 | f | 1–51 | – | 52 |
| eL41 | C6S3G4_PLAF7 | PF3D7_1144300 | g | 3–39 | 1–14 | 39 |
| eL43 | RL37A_PLAF7 | PF3D7_0210100.1 | h | 2–86 | – | 96 |
| eL44 | RL44_PLAF7 | PF3D7_0304400 | i | 2–96 | – | 104 |
Comparison of ESs in Pf80S and human cytoplasmic ribosomes
| rRNA | ES | Helix | Comparison between Pf80S and human ribosomes |
|---|---|---|---|
| 18S | ES2S | Shorter loop in Pf80S | |
| ES3S | A | Conserved | |
| B | Truncated in Pf80S | ||
| ES13S | Conserved | ||
| ES6S | A | Expanded in Pf80S | |
| B | Truncated in Pf80S | ||
| C | Conserved | ||
| D | Expanded in Pf80S | ||
| E | Conserved | ||
| ES7S | Expanded in Pf80S | ||
| ES14S | Conserved | ||
| ES9S | Expanded in Pf80S | ||
| ES10S | Expanded in Pf80S | ||
| ES12S | Helix truncated in Pf80S | ||
| 28S | ES3L | Conserved | |
| ES4L | Conserved | ||
| ES5L | Conserved | ||
| ES7L | A | Truncated in Pf80S | |
| B | Truncated. Loop in Pf80S forms a novel interaction with eL14 | ||
| B1 | Pf-specific ES | ||
| C | Present | ||
| D–H | Absent from Pf80S | ||
| ES8L | H28 | Expanded in Pf80S | |
| ES9L | A | Absent in Pf80S | |
| H30 | Conserved | ||
| H31 | Conserved | ||
| ES10L | Absent in Pf80S | ||
| ES12L | Expanded in Pf80S | ||
| ES15L | A | Truncated in Pf80S | |
| ES19L | Truncated in Pf80S | ||
| ES20L | A | Absent in Pf80S | |
| B | Conserved in Pf80S | ||
| ES26L | Expanded in Pf80S | ||
| ES27L | A–C | Not present in Pf80S model, predicted divergence between Pf and human cytoplasmic ribosomes | |
| ES30L | Absent in Pf80S | ||
| ES31L | A | Conserved | |
| B | Expanded in Pf80S | ||
| C | Conserved | ||
| ES34L | Pf-specific ES | ||
| ES36L | Pf-specific ES | ||
| ES39L | A | Conserved; preceding loop in Pf80S forms a short helix (3 base pairs) with the 5′ end of the 5.8S rRNA | |
| B | Conserved | ||
| ES41L | Conserved |





