Analysis of the crystal structure of an active MCM hexamer
Figures
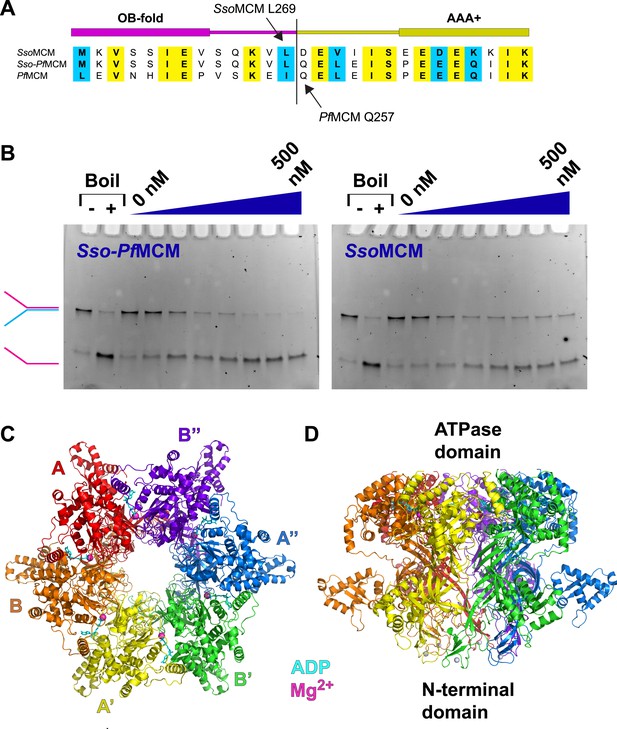
Properties of Sso-PfMCM.
(A) Sequence alignment showing the construction of the Sso-PfMCM chimera. The N-terminal domain of SsoMCM (residues 1–269, top sequence) was fused to the AAA+ domain of PfMCM (starting at residue 257, bottom sequence) to yield the chimera (middle sequence). (B) The Sso-PfMCM chimera shows enhanced unwinding activity when compared to wild-type SsoMCM. Helicase reactions were performed at 69°C for 60 min with a Y-shaped DNA substrate with a 5ʹ-fluorescein label on one strand. Unwinding reactions were in the presence of 4 mM ATP and contained 0, 50, 100, 150, 200, 300, 400, or 500 nM protein. Views of the Sso-PfMCM hexamer crystal structure parallel (C) and perpendicular (D) to the central channel with each subunit uniquely colored. The magnesium ions are magenta spheres, and ADP molecules are shown as cyan stick. (C) View down the crystallographic threefold axis with the unique and symmetry-derived chains labeled. The ATPase domains are projected out of the page. (D) View perpendicular to the channel axis. The ATPase domains are located at the top, and the N-terminal domains are located at the bottom. The Zn ions are light grey spheres at the bottom.
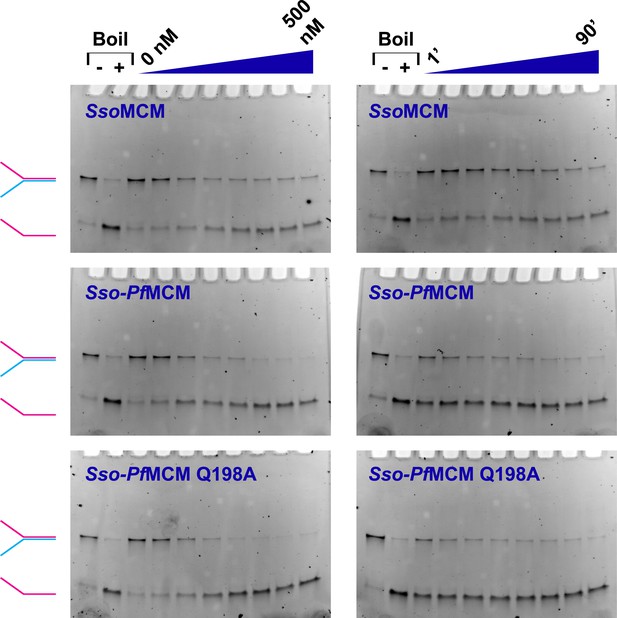
MCM catalyzed DNA unwinding visualized by gel electrophoresis.
MCM unwinding activity was observed as a function of protein concentration (left column; 0, 50, 100, 150, 200, 300, 400, and 500 nM protein) or time (right column; 1, 5, 10, 20, 30, 40, 60, and 90 min). A Y-shaped substrate (3.7 nM) was incubated at 69°C with the protein and 4 mM ATP. Time courses were collected by quenching independent samples (final composition of quench solution is 8% (vol/vol) glycerol, 1% (wt/vol) SDS, and 10 mM EDTA). Lanes marked with ‘−’ or ‘+’ are control samples without reaction (‘−’) or boiled at 98°C (‘+’), respectively. At left, the positions of intact Y-shaped substrate (red and blue) and displaced ssDNA (red) are indicated. The Sso-PfMCM chimera shows an enhanced unwinding acivity relative to SsoMCM, and the Sso-PfMCM Q198A mutant shows no apparent defect in unwinding.
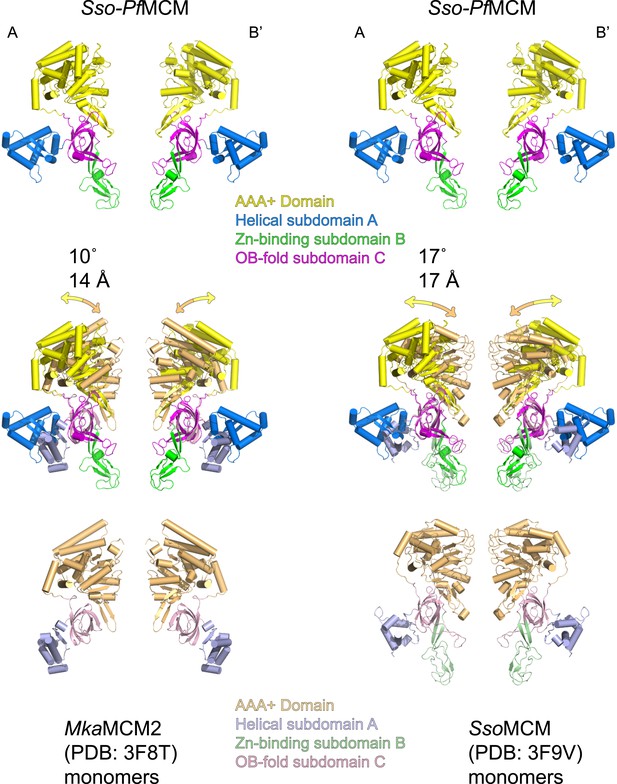
The relative positions of the N- and C-terminal domains in Sso-PfMCM significantly differ from previous monomeric crystal structures.
Two monomeric MCM crystal structures (3F8T, Bae et al., 2009, left; and 3F9V, Brewster et al., 2008, right) were superimposed on two subunits of the Sso-PfMCM hexamer (at opposite sides of the channel) crystal structure based on the OB-fold subdomain. For the corresponding AAA+ domain positions, the differences in rotation and center-of-mass position were calculated and expressed in degrees and Å (middle). The AAA+ domains of Sso-PfMCM are rotated further from the N-terminal domain, which is necessary to prevent clashes that would occur among the six AAA+ domains if six copies of a monomeric structure were superimposed on each OB-fold of the Sso-PfMCM hexamer.
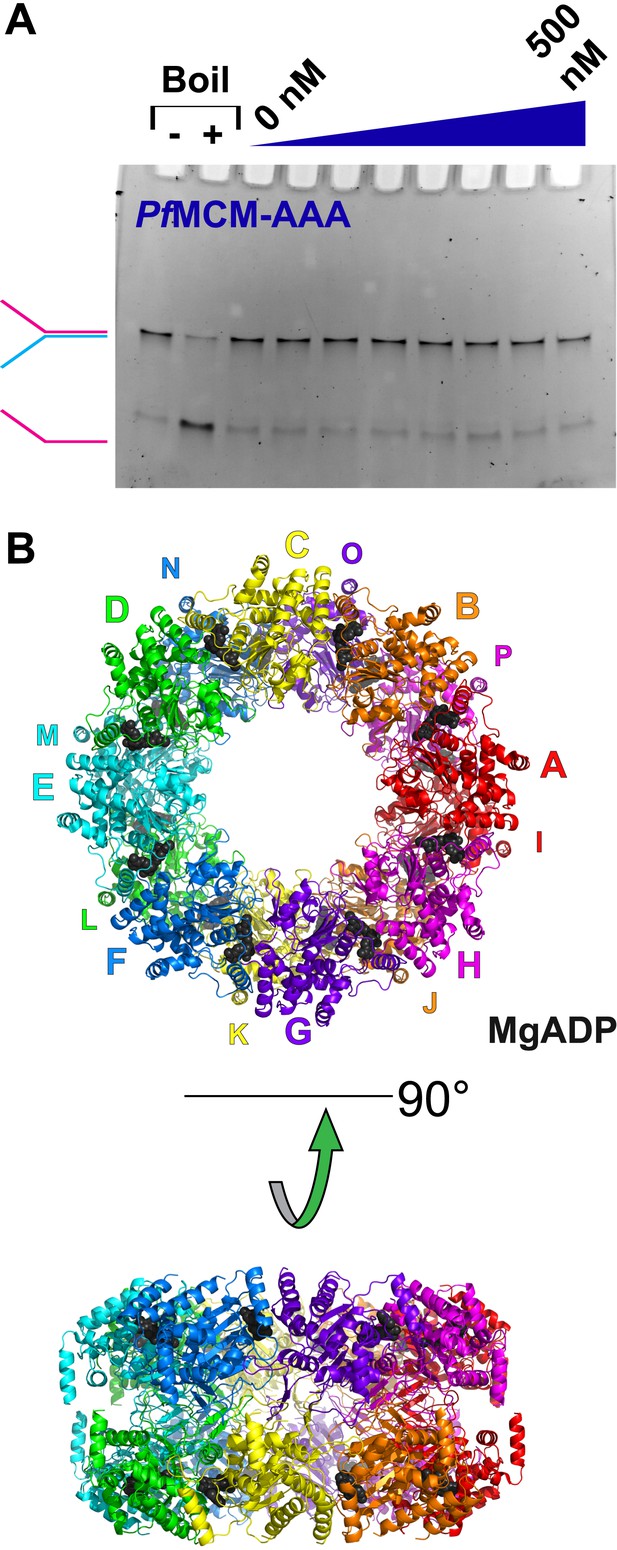
Activity and structure of PfMCMAAA.
(A) An unwinding experiment with varying protein concentrations analogous to Figure 1 (0, 50, 100, 150, 200, 300, 400, and 500 nM protein) showed negligible unwinding. (B) The crystal structure of PfMCMAAA (see ‘Materials and methods’) reveals a double-octamer architecture and an unusual topology for the h2i to mediate the interface between the octamers. The unusual h2i topology is not observed the Sso-PfMCM hexamer. A non-hexameric architecture or an unusual h2i topology could contribute to the inactivity of this domain.
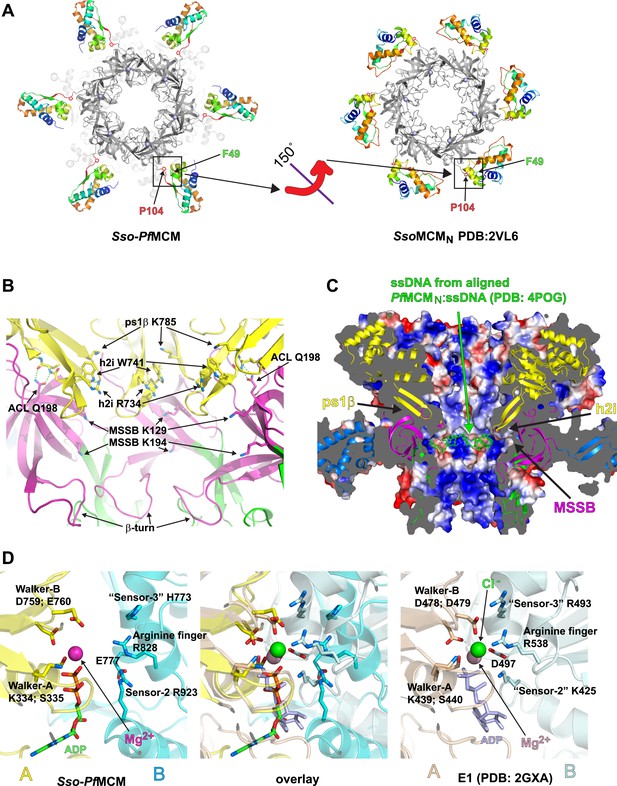
Sso-PfMCM crystal structure details.
(A) The A-subdomains (rainbow) of Sso-PfMCM adopt a unique conformation that is rotated 150° compared to other crystal structures of hexameric MCMN such as SsoMCMN (Liu et al., 2008). The distinct conformations correlate with the conformation of P104 that is located at the junction between the A- and C-subdomains (boxed). In both conformations, P104 packs against the aromatic residue F49. (B) The modules of the central channel. The cartoon is colored with the AAA+ domain in yellow, the Zn-binding B-subdomain in green, and the OB-fold C-subdomain in magenta. The ps1β projects a conserved lysine, K785, into the channel. This lysine packs against W741 of the h2i, which sits adjacent to R734 of the h2i. The MSSB is recessed and sits below the h2i and above the β-turn. The N-terminal domain is tethered to the h2i by a universally conserved glutamine in the ACL, Q198. (C) Surface representation of Sso-PfMCM colored by electrostatic potential. The surface is clipped with a vertical plane through the center to illustrate the central channel features. A cartoon representation of the protein with select modules labeled is colored as in Figure 2B with the helical A-subdomain in blue. The MSSB sits at a recessed pocket. The ssDNA from the aligned PfMCMN:ssDNA structure (Froelich et al., 2014) would be snugly positioned in this pocket. (D) Comparison of the Sso-PfMCM ATPase site (left) with that of E1 (Enemark and Joshua-Tor, 2006, right). The Walker-A and Walker-B residues of one subunit (yellow) are positioned at the left side of the site while three positively charged residues of the adjacent subunit (cyan) line the right side of the site. An acidic residue of the cyan subunit sits below the site. Based on the superposition (middle), we predict that the MCM subunits need to approach each other more closely to generate a competent ATPase site.
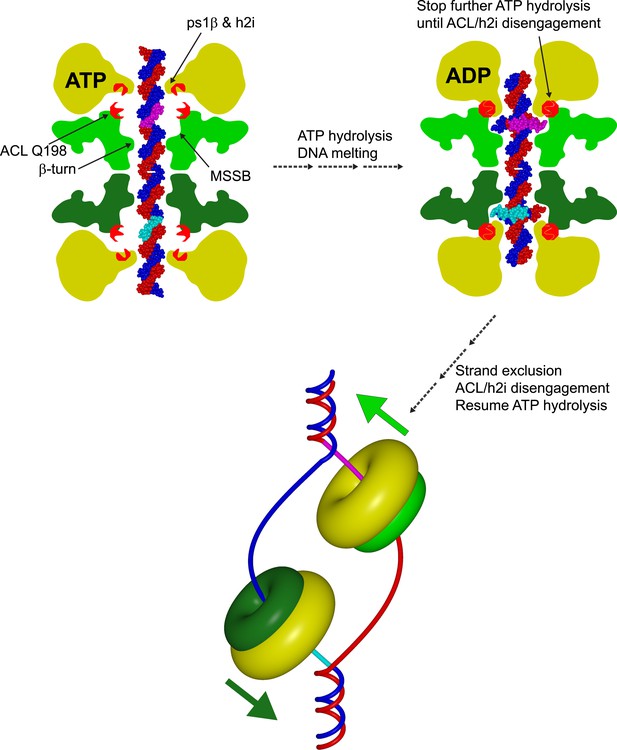
Cartoon model showing a role for an interaction between the N-terminal ACL conserved glutamine and the AAA+ h2i during helicase activation.
The N-terminal domains of each hexamer are represented in different shades of green, and the AAA+ domains are represented in yellow. A double-hexamer initially encircles dsDNA (blue and red strands). In the ATP-bound state (left), the ps1β and h2i modules are located further from the ACL than in the ADP state (right) as identified previously for the ACL/ps1β of SsoMCM (Barry et al., 2009). ATP-hydrolysis could drive the h2i/ps1β inward to generate strand separation that is captured by the MSSB as shown in Figure 7 of (Froelich et al., 2014). In this position, ACL Q198 is able to interact with the h2i, clamping down on the bound portion of ssDNA (cyan and magenta portions of ssDNA). ATP hydrolysis stops until the h2i is able to return to the starting ‘ATP’ position, potentially after an important activation criteria is reached. This species is ultimately converted to a strand-excluded complex able to unwind DNA (bottom).
Videos
Crystal structure details for Sso-PfMCM.
The video illustrates the arrangement of the subunits in the hexamer and the positions of the subdomains. The A-subdomain conformation is animated to transform to that observed in other crystal structures of MCMN to illustrate how they differ. The different A-subdomain conformations correlate with the conformation of a proline (P104) at the junction between the A- and C-subdomains. The relative position of the mcm5-bob1 mutation is noted. Several central channel modules are highlighted, including the ps1β, h2i, β-turn, MSSB, and the interaction of ACL Q198 with the h2i. The ATPase site is compared to that of papillomavirus E1 (Enemark and Joshua-Tor, 2006), and several key residues are highlighted for MCM. The MSSB location is shown in a surface representation to illustrate that it sits at a recessed binding pocket where the ssDNA (green) of the aligned PfMCMN:ssDNA crystal structure (Froelich et al., 2014) would position snugly.
Tables
Data collection and refinement statistics
| Sso-PfMCM:MgADP Hexamer | PfMCMAAA:MgADP Double-octamer | |
|---|---|---|
| Data collection | ||
| Space group | P63 | P1 |
| Cell dimensions | ||
| a, b, c (Å) | 118.902, 118.902, 199.317 | 124.956, 127.082, 128.025 |
| α, β, γ (°) | 90, 90, 120 | 71.852, 72.819, 80.392 |
| Resolution (Å) | 50–2.70 (2.80–2.70) | 50–3.80 (3.94–3.80) |
| Rsym | 0.107 (0.750) | 0.169 (0.429) |
| I/σI | 14.8 (1.79) | 8.3 (2.41) |
| Completeness (%) | 99.8 (98.3) | 98.9 (97.0) |
| Redundancy | 6.8 (5.0) | 3.3 (2.6) |
| Refinement | ||
| Resolution (Å) | 50–2.70 (2.80–2.70) | 50–3.80 (3.94–3.80) |
| No. reflections | 39,044/1976 (2042/123) | 69,126/3486 (6206/357) |
| Rwork/Rfree | 0.263/0.295 (0.360/0.353) | 0.301/0.314 (0.367/0.368) |
| No. atoms | ||
| Protein | 9432 | 2429 (1/16 of ASU) |
| ADP | 54 | 27 (1/16 of ASU) |
| ions | 10 | 1 (1/16 of ASU) |
| Water | 0 | 0 |
| B-factors | ||
| Protein | 60 | 91 |
| ADP | 118 | 72 |
| ions | 75 | 81 |
| Water | N/A | N/A |
| R.m.s. deviations | ||
| Bond lengths (Å) | 0.010 | 0.010 |
| Bond angles (°) | 1.488 | 1.597 |
Additional files
-
Supplementary file 1
ClustalW sequence alignment of several MCM proteins in MSF format. Portions of this alignment are shown throughout Video 1. The Sso-PfMCM residue numbers in the alignment run consecutively, which is consistent with database numbering for the SsoMCM portion. For the PfMCM portion, database numbers can be obtained from the aligned PfMCM sequence. The intein (residues 362–728) was manually removed from the PfMCM sequence for the alignment. Thus, for residues of PfMCM in the alignment with sequence numbers greater than 361, 367 must be added to agree with database (intein-containing) sequence numbering. Residue numbering throughout the manuscript and Video 1 agrees with the database numbering for the respective portions.
- https://doi.org/10.7554/eLife.03433.010





