Redox signaling via the molecular chaperone BiP protects cells against endoplasmic reticulum-derived oxidative stress
Figures
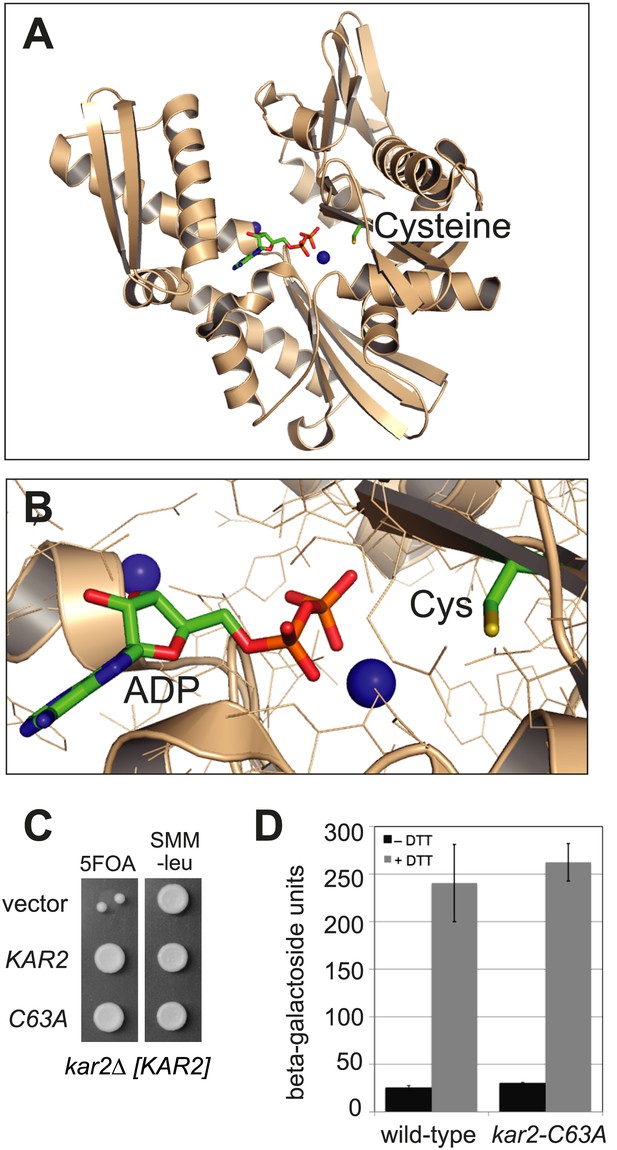
BiP contains a conserved cysteine that is dispensable for yeast viability.
(A) Ribbon diagram of human BiP nucleotide binding domain in complex with calcium-ADP (PDB entry 3IUC) (Wisniewska et al., 2010). ADP and the conserved BiP cysteine are shown as colored sticks. Calcium atoms are represented as blue spheres. (B) Magnified representation of the conserved cysteine and CaADP from the BiP structure in panel A. Additional amino acid side chains are shown as lines. (C) CSY214 containing the plasmids pCS681, pCS685, or empty vector were spotted onto SMM plates with or without 5-fluoroorotic acid (5-FOA) and incubated for 2 d at 30°C. (D) CSY5 or CSY275 containing a UPRE-lacZ reporter plasmid (pJC8) were cultured in SMM-ura at 30°C, treated with 0 or 2 mM dithiothreitol (DTT) for 2 hr, and assayed for beta–galactosidase activity. Three independent transformants of each strain were grown and assayed in duplicate. Data represent the mean of averaged values for the three transformants ± SD.
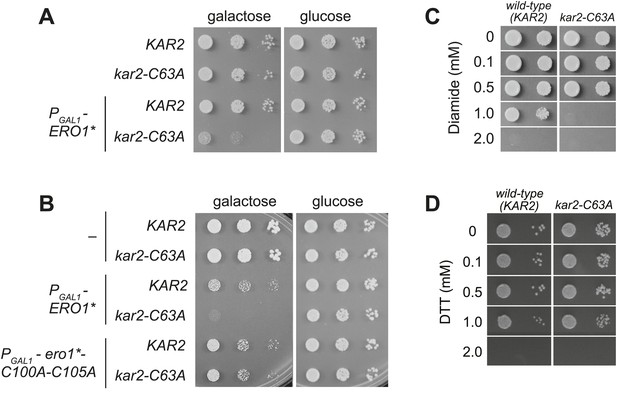
A cysteine-less BiP strain is sensitive to increased ER oxidation.
(A) CSY170 and CSY278 strains containing an integrated galactose-inducible ERO1* were spotted onto SMM or SMM Gal plates, and plates were incubated for 2 d (glucose) or 3 d (galactose) at 30°C. (B) CSY5 or CSY275 strains transformed with plasmids pCS452, pCS504, or empty vector were spotted onto SMM-ura or SMM Gal-ura plates and incubated for 2 d (glucose) or 3 d (galactose) at 30°C. (C and D) CSY5 or CSY275 strains were spotted onto SMM plates containing 0–2 mM diamide (C) or DTT (D), and plates were incubated at 30°C for 2 d.
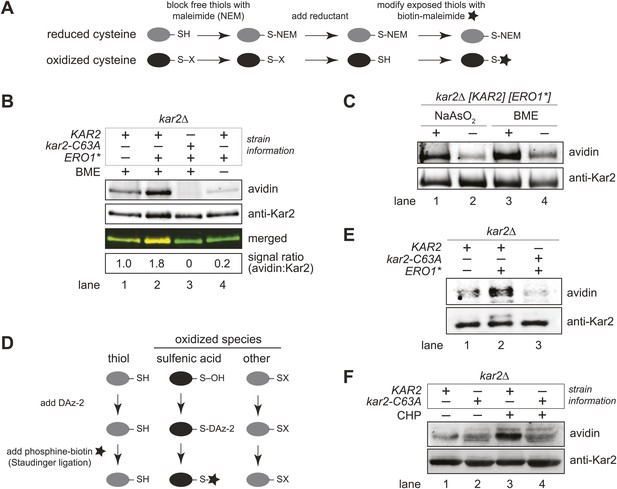
BiP's cysteine is oxidized upon hyperoxidation of the ER by Ero1*.
(A) Schematic for the biotin-switch assay used in panels B and C. (B) The biotin-switch assay was used on lysates prepared from strains CSY316 and CSY319 containing either pCS452 or an empty vector. Strains were grown in galactose medium to induce Ero1*, and Kar2 was immunoprecipitated from lysates postbiotin-maleimide treatment. The relative proportion of Kar2 with an oxidized cysteine in the cell lysates under stressed (ERO1*) and non-stressed conditions was determined by comparing the intensity of the Kar2-biotin signal relative to the total level of Kar2. As a control (lane 4), no reductant was added postNEM treatment and prior to biotin-maleimide addition. (C) Lysates were prepared from CSY316 containing pCS452 grown in galactose medium. The biotin-switch assay was performed with BME (as in panel B) or sodium arsenite (NaAsO2) as the reductant. Kar2 was immunoprecipitated from lysates postbiotin-maleimide treatment. (D) Schematic for the use of DAz-2 to detect sulfenic acid in panel E and F. (E) Strains were grown as in panel B and treated with DAz-2. Kar2 was immunoprecipitated from lysates, and Staudinger ligation was performed with phosphine-biotin. DAz-2 addition was detected with an avidin probe. Note, a higher molecular weight Kar2 band corresponding to untranslocated Kar2 was observed in the Ero1*-overexpression strain (lane 2). Overlay of the two images confirmed that the avidin signal corresponds to the lower molecular weight mature Kar2 band (data not shown). (F) Strains were grown in glucose medium, and cells were exposed to 5 mM cumene hydroperoxide (CHP) for 30 min prior to harvest. Treatment with DAz-2 and sample processing were as described in panel E.
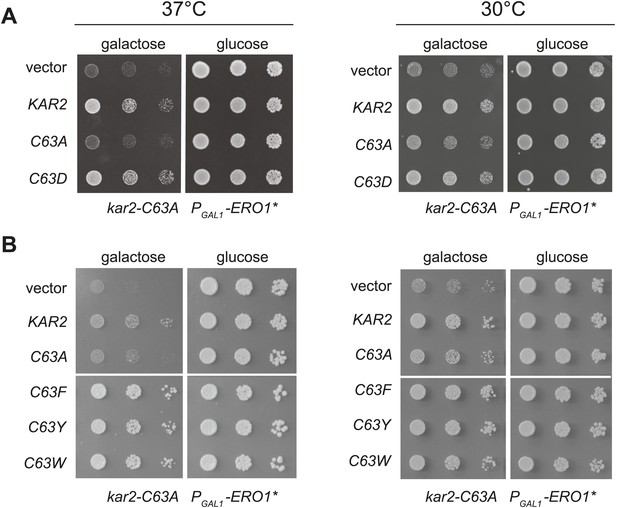
Substitution of the BiP cysteine with an amino acid containing a negatively charged or large side chain enables protection against hyper-oxidation of the ER lumen.
(A and B) CSY278 containing (A) plasmids pCS681, pCS685, pCS802, or empty vector and (B) plasmids pCS681, pCS685, pCS687, pCS688, pCS750, or empty vector were spotted on SMM-leu or SMM Gal-leu plates, and plates were incubated at 30°C and 37°C for 2 d (glucose) or 3 d (galactose).
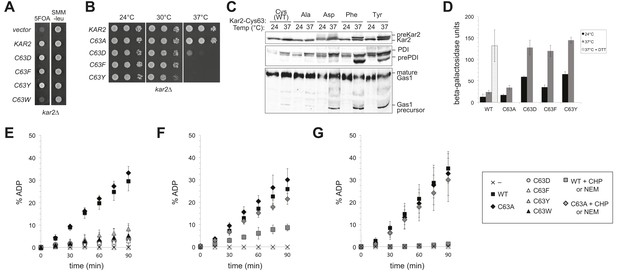
Replacement of the BiP cysteine with aspartic acid, phenylalanine, tyrosine, or tryptophan results in decreased BiP function.
(A) CSY214 containing the plasmids pCS681, pCS802, pCS687, pCS688, pCS750 or empty vector were spotted onto SMM plates with or without 5-fluoroorotic acid (5-FOA) and incubated for 2 d at 30°C. (B) CSY289, 290, 368, 292, and 293 were spotted onto YPD plates and incubated at 24°, 30°, and 37°C. (C) Strains from B were cultured at 24°C to log-phase in YPD and shifted to 37°C for 90 min prior to harvest. Accumulation of unprocessed untranslocated forms of the proteins Kar2, PDI, and Gas1 were detected by western blotting. (D) Strains from B containing an UPRE-lacZ reporter plasmid (pJC8) were cultured in SMM-ura at 24°C to log-phase and shifted to 37°C (with or without 2 mM DTT) for 90 min prior to harvest. Samples were assayed for beta–galactosidase activity. Three independent transformants of each strain were grown and assayed in duplicate. Data represent the mean of averaged values for the three transformants ± SD. (E–G) ATP hydrolysis was assessed by determining the fraction of [alpha-32P]ATP converted to [alpha-32P]ADP as described in the 'Materials and methods'. Panels F and G show CHP and NEM-treated samples, respectively. Samples without chemical additions in panels F and G were mock treated to match the CHP or NEM-treatment. Data represent the means ± SD of three independent assays.
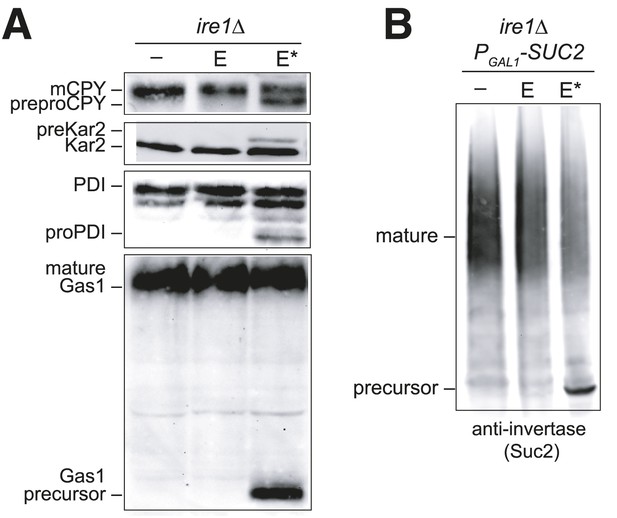
Overexpression of Ero1* causes an accumulation of untranslocated polypeptides.
(A) CSY44 and (B) CSY172 containing plasmids pAF112 (ERO1; E), pCS452 (ERO1*; E*), or empty vector were cultured in galactose medium for 5 hr to induce Ero1 and Suc2 expression. Accumulation of unprocessed untranslocated forms of the proteins CPY, Kar2, PDI, Gas1, and Suc2 were detected by western blotting.
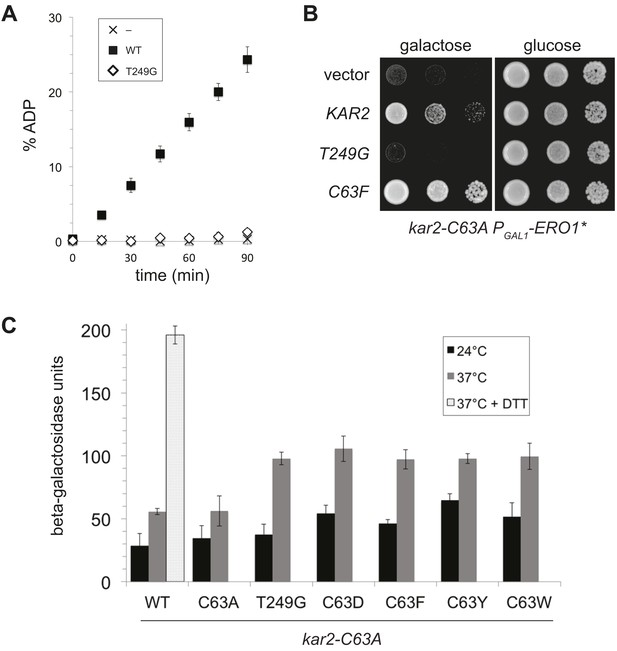
A BiP ATPase mutant is not sufficient to protect cells during oxidative stress.
(A) ATP hydrolysis was assessed by determining the fraction of [alpha-32P]ATP converted to [alpha-32P]ADP as described in the 'Materials and methods'. Data represent the means ± SD of three independent assays. (B) CSY278 containing plasmids pCS681, pCS774, pCS687, or empty vector were spotted on SMM-leu or SMM Gal-leu plates, and plates were incubated at 37°C for 3 d. (C) CSY275 containing a UPRE-lacZ reporter (pCS852) and plasmids pCS681, pCS802, pCS774, pCS687, pCS688, or pCS750 were cultured in SMM-ura-leu at 24°C to log-phase and shifted to 37°C (with or without 2 mM DTT) for 90 min prior to harvest. Three independent transformants of each strain were grown and assayed for beta–galactosidase activity in duplicate. Data represent the mean of averaged values for the three transformants ± SD.
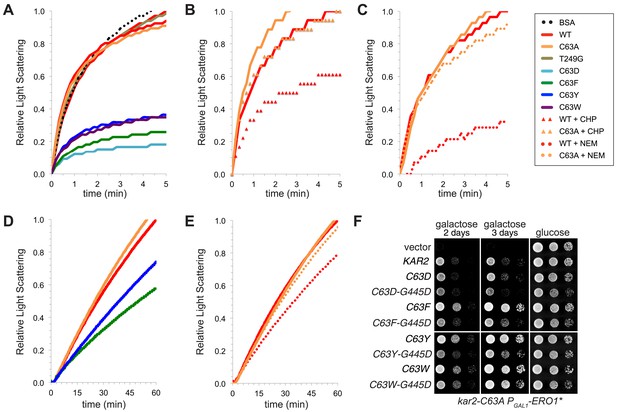
BiP cysteine mutants that protect cells during oxidative stress are more effective than wild-type BiP in suppressing polypeptide aggregation.
(A–C) Denatured rhodanese was diluted to a final concentration of 1 µM in the presence of 4 µM BSA or wild-type, mutant, peroxide-treated, or alkylated BiP. Samples in panels B and C were mock treated to match the CHP or NEM-treatment. Rhodanese aggregation was followed by monitoring light scattering at 320 nm over a period of 5 min (D and E) After denaturation in 6 M guanidine and 40 mM DTT, IgY was diluted to final concentration of 0.7 µM at 45°C in the presence or absence of 0.7 µM wild-type, mutant, or alkylated BiP. IgY aggregation was followed by monitoring light scattering at 360 nm over a period of 60 min. Data in panels A–E are representative traces from at least three trials. (F) CSY278 containing plasmids pCS681, pCS802, pJW5, pCS687, pCS844, pCS688, pCS845, pCS750, pCS846, or empty vector were spotted on SMM-leu or SMM Gal-leu plates, and plates were incubated at 37°C for 2 d (glucose) or 2-3 d (galactose).
Tables
Plasmids
| Name | Description | Markers | Source |
|---|---|---|---|
| pJC8 | UPRE-LacZ reporter | CEN URA3 LEU2 | Cuozzo and Kaiser, 1999 |
| pCS852 | UPRE-LacZ reporter | CEN URA3 | This study |
| pAF112 | PGAL1-ERO1-myc | CEN URA3 | Sevier et al., 2007 |
| pCS452 | PGAL1-ERO1*-myc | CEN URA3 | Sevier et al., 2007 |
| pCS504 | PGAL1-ero1*-C100A-C105A-myc | CEN URA3 | Sevier et al., 2007 |
| pCS584 | can1::PGAL1-ERO1*-myc | CEN URA3 | This study |
| pCS739 | kar2-C63A | URA3 | This study |
| pCS623 | KAR2 | CEN URA3 | This study |
| pCS681 | KAR2 | CEN LEU2 | This study |
| pCS685 | kar2-C63A | CEN LEU2 | This study |
| pCS802 | kar2-C63D | CEN LEU2 | This study |
| pCS687 | kar2-C63F | CEN LEU2 | This study |
| pCS688 | kar2-C63Y | CEN LEU2 | This study |
| pCS750 | kar2-C63W | CEN LEU2 | This study |
| pCS774 | kar2-T249G | CEN LEU2 | This study |
| pJW5 | kar2-C63D-G445D | CEN LEU2 | This study |
| pCS844 | kar2-C63F-G445D | CEN LEU2 | This study |
| pCS845 | kar2-C63Y-G445D | CEN LEU2 | This study |
| pCS846 | kar2-C63W-G445D | CEN LEU2 | This study |
| pCS757 | KAR2-FLAG | CEN LEU2 | This study |
| pCS760 | kar2-C63A-FLAG | CEN LEU2 | This study |
| pCS630 | kar2-(40-668)-His6 | AMP | This study |
| pCS631 | kar2-(40-668)-C63A-His6 | AMP | This study |
| pJW4 | kar2-(40-668)-C63D-His6 | AMP | This study |
| pCS658 | kar2-(40-668)-C63F-His6 | AMP | This study |
| pCS643 | kar2-(40-668)-C63Y-His6 | AMP | This study |
| pCS644 | kar2-(40-668)-C63W-His6 | AMP | This study |
| pCS639 | kar2-(40-668)-T249G-His6 | AMP | This study |
| pCS675 | GST-sec63J-(121-221) | AMP | This study |
| pCS817 | His6-kar2-(42-682) | KAN | This study |
| pCS818 | His6-kar2-(42-682)-C63A | KAN | This study |
| pCS822 | His6-kar2-(42-682)-C63D | KAN | This study |
| pCS819 | His6-kar2-(42-682)-C63F | KAN | This study |
| pCS820 | His6-kar2-(42-682)-C63Y | KAN | This study |
| pCS821 | His6-kar2-(42-682)-C63W | KAN | This study |
| pKP113 | His6-kar2-(42-682)-T249G | KAN | This study |
Yeast strains
| Strain | Genotype | Source |
|---|---|---|
| CKY263/CSY5 | MATa GAL2 ura3-52 leu2-3,112 | Lab collections |
| CKY264/CSY6 | MATα GAL2 ura3-52 leu2-3,112 | Lab collections |
| CKY1026/CSY44 | MATa GAL2 ura3-52 leu2-3,112 ire1Δ::KanMX | Lab collections |
| CSY158 | MATa GAL2 ura3-52 leu2-3,112 ire1Δ::NatMX | This study |
| CSY172 | MATa GAL2 ura3-52 leu2-3,112 ire1Δ::NatMX KanMX:PGAL1-SUC2 | This study |
| CSY275 | MATa GAL2 ura3-52 leu2-3,112 kar2-C63A | This study |
| CSY277 | MATa GAL2 ura3-52 leu2-3,112 kar2-C63A ire1Δ::NatMX | This study |
| CSY170 | MATa GAL2 ura3-52 leu2-3,112 can1::PGAL1-ERO1*-myc | This study |
| CSY278 | MATa GAL2 ura3-52 leu2-3,112 kar2-C63A can1::PGAL1-ERO1*-myc | This study |
| CSY214 | MATa GAL2 ura3-52 leu2-3,112 kar2Δ::KanMX [pCS623] | This study |
| CSY289 | MATa GAL2 ura3-52 leu2-3,112 kar2Δ::KanMX [pCS681] | This study |
| CSY290 | MATa GAL2 ura3-52 leu2-3,112 kar2Δ::KanMX [pCS685] | This study |
| CSY368 | MATa GAL2 ura3-52 leu2-3,112 kar2Δ::KanMX [pCS802] | This study |
| CSY292 | MATa GAL2 ura3-52 leu2-3,112 kar2Δ::KanMX [pCS687] | This study |
| CSY293 | MATa GAL2 ura3-52 leu2-3,112 kar2Δ::KanMX [pCS688] | This study |
| CSY308 | MATa GAL2 ura3-52 leu2-3,112 kar2Δ::KanMX pep4Δ::NatMX [pCS623] | This study |
| CSY316 | MATa GAL2 ura3-52 leu2-3,112 kar2Δ::KanMX pep4Δ::NatMX [pCS757] | This study |
| CSY319 | MATa GAL2 ura3-52 leu2-3,112 kar2Δ::KanMX pep4Δ::NatMX [pCS760] | This study |





