The Drosophila Sp8 transcription factor Buttonhead prevents premature differentiation of intermediate neural progenitors
Figures
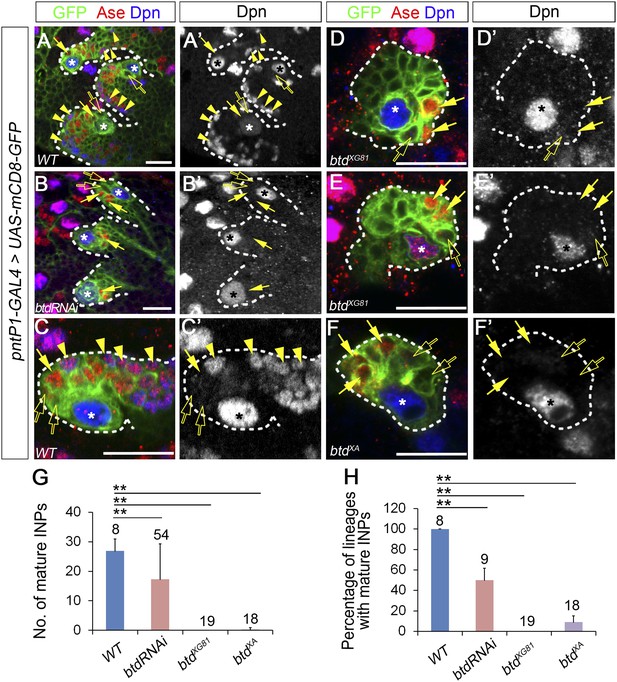
Loss of Btd eliminates mature INPs in type II NB lineages.
(A–A′, C–C′) Wild-type type II NB lineages in a third instar larval brain. mCD8-GFP driven by pntP1-GAL4 labels all type II NB lineages (A–A′) or a single type II NB clone (C–C′). Ase− immature INPs, Ase+ immature INPs, and mature INPs are indicated by open arrows, solid arrows, and arrowheads, respectively. (B–B′, D–F′) Btd RNAi knockdown type II NB lineages (B–B′) or type II NB clones homozygous mutant for btdXG81 (D–E′) or btdXA (F–F′) in 3rd instar larval brains produce Ase− immature INPs (open arrows) and a few Ase+ daughter cells (arrows) but no mature INPs. Only 3 out of total 8 type II NB lineages are shown in (A–A′) and (B–B′). In this and all other figures, asterisks indicate type II NBs and scale bars equal to 20 µm. Dpn staining alone shows the NB and mature INPs. (G–H) Quantifications of the number of mature INPs (G) and the percentage of type II NB lineages with mature INPs (H) in the wild type, Btd RNAi knockdown, and btd mutant type II NB lineages. The numbers on top of each bar are the numbers of type II NB lineages analyzed except for the numbers for the wt and btd RNAi in (H), which are the number of brain lobes examined. The mean and stdev for btdXG81 and btdXA in (H) are calculated by bootstrapping. **p < 0.01, *p < 0.05 (Student t test).
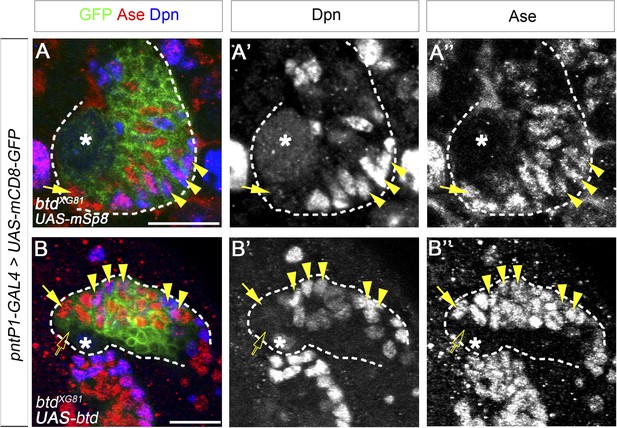
Expression of mouse Sp8 (mSp8).
(A–A′′) and Drosophila Btd (B–B′′) rescues the loss of mature INPs in btd mutant type II NB clones. Multiple mature INPs are observed in btd mutant type II NB clone that expresses UAS-mSp8 (A–A′′) or UAS-btd (B–B′′). Type II NBs, Ase− immature INPs, Ase+ immature INPs, and mature INPs are indicated by asterisks, open arrows, solid arrows, and arrowheads, respectively.
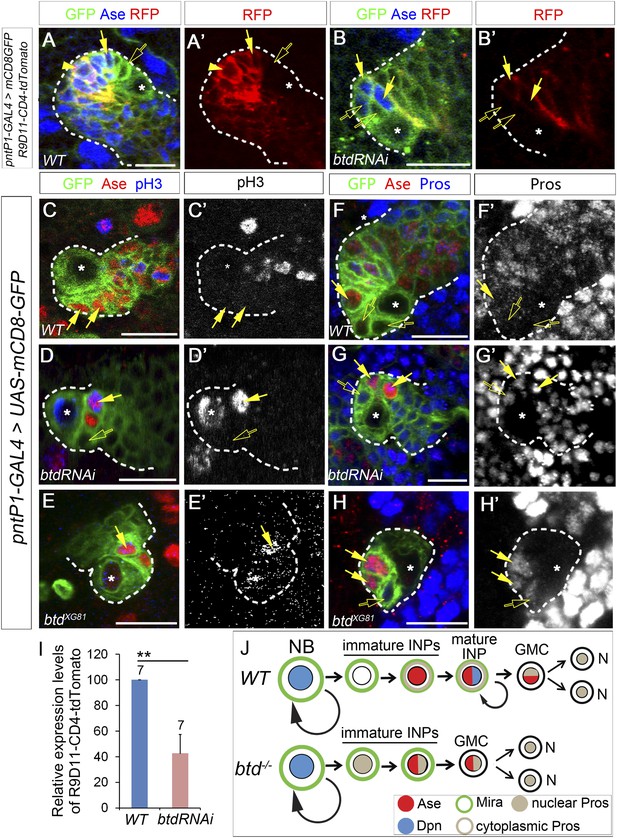
Loss of Btd results in ectopic nuclear Pros in immature INPs and premature differentiation of Ase+ immature INPs into GMCs.
(A–A′) R9D11-CD4-tdTomato is expressed in Ase+ immature INPs (solid arrows) and mature INPs (arrowheads) but not in Ase− immature INPs (open arrows) in a wild-type type II NB lineage. (B–B′) R9D11-CD4-tdTomato remains expressed in Ase+ daughter cells (solide arrows) next to the Ase− immature INPs (open arrows) when mature INPs are eliminated by Btd RNAi knockdown. (C–C′) pH3 is not detected in Ase+ immature INPs (solid arrows) in a wild-type type II NB lineage. (D–E′) pH3 is expressed in Ase+ daughter cells (solid arrows) that are the furthest from the Ase− immature INPs (open arrows) in a Btd RNAi knockdown type II NB lineage without mature INPs (D–D′) or btd mutant type II NB lineages (E–E′). (F–F′) Nuclear Pros is not expressed in Ase− immature INPs (open arrows) in a wild-type type II NB lineage. (G-H′) Nuclear Pros is ectopically expressed in both Ase− immature INPs (open arrows) and Ase+ cells (solid arrows) in Btd RNAi knockdown (G–G′) or btd mutant (H–H′) type II NB lineages. (I) Quantifications of relative overall expression levels of R9D11-CD4-tdTomato in wild-type (A–A′) and Btd RNAi knockdown (B–B′) type II NB lineages. (J) A diagram of neurogenesis patterns in type II NB lineages in the presence or absence of Btd.
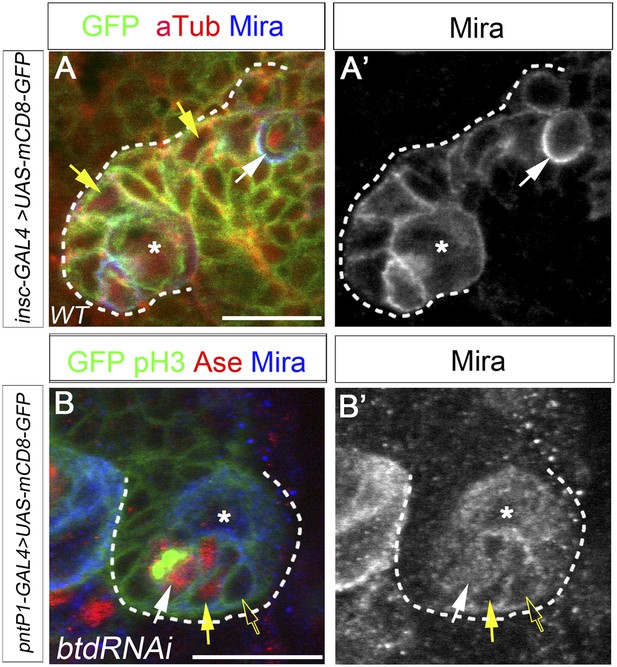
Mira expression in Btd RNAi knockdown type II NB clone.
(A–A′) Mira is expressed in INPs (yellow arrows) in a wild-type type II NB lineage and forms a basal crescent (white arrows) at metaphase. (B–B′) In a Btd RNAi knockdown type II NB lineage without mature INPs, Mira is expressed in Ase+ daughter cells (yellow arrows) next to the Ase− immature INP (open arrows) but does not form a basal crescent at metaphase in the dividing Ase+ cell (white arrows).
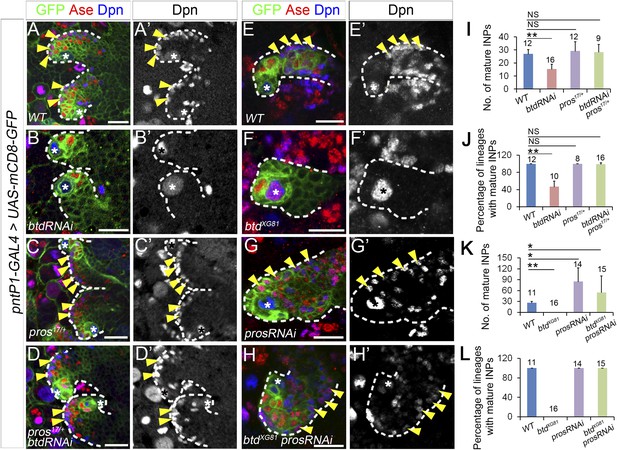
Reducing Pros rescues the elimination of INPs resulting from the loss of Btd.
(A–D′) The loss of INPs resulting from Btd RNAi knockdown is rescued in pros17 heterozygous mutant background. Only two lineages are shown in each brain. (A–A′) Wild-type type II NB lineages have multiple mature INPs. (B–B′) Btd RNAi knockdown causes a loss of mature INPs. (C–C′) Type II NB lineages in pros17 heterozygous mutant larvae produce a similar number of mature INPs as in wild-type larvae. (D–D′) Btd RNAi knockdown no long leads to the loss of mature INPs in pros17 heterozygous mutant type II NB lineages. (E–H′) Pros RNAi knockdown rescues the loss of INPs in btd mutant type II NB clones. (E–E′) A wild-type type II NB clone has multiple mature INPs. (F–F′) A btd mutant type II NB clone contains no mature INPs. (G–G′) Pros RNAi knockdown causes overproliferation of mature INPs in a type II NB clone. (H–H′) Pros RNAi knockdown rescues the loss of mature INPs in a btd mutant type II clone. Arrowheads point to mature INPs in all images. (I–L) Quantifications of the number of mature INPs (I–K) and the percentage of lineages with mature INPs (J–L) for the rescue of Btd RNAi knockdown phenotypes in pros17/+ larvae (I–J) or the rescue of btd mutant phenotypes by Pros RNAi knockdown (K–L). The samples sizes on top of each bar represent the number of type II NB lineages (I, K, L) or the number of brain lobes (J). The mean and stdev in (L) are calculated by bootstrapping. **p < 0.01, *p < 0.05 (Student t test). NS: not significant.
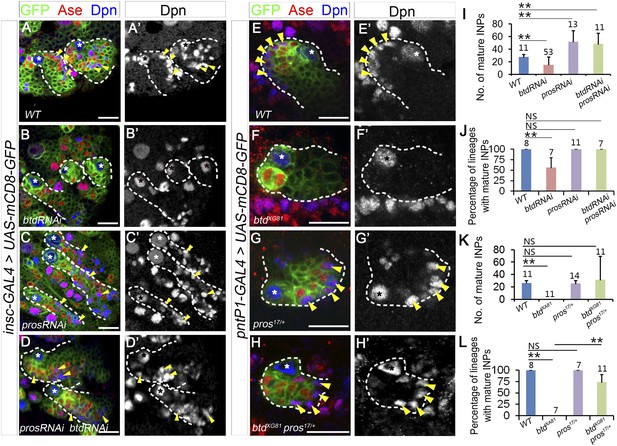
Reducing Pros rescues the elimination of INPs resulting from the loss of Btd.
(A–D′) The loss of INPs resulting from Btd RNAi knockdown is rescued by Pros RNAi knockdown. (A–A′) Wide-type type II lineages labeled with mCD8-GFP driven by insc-GAL4 have multiple mature INPs. (B–B′) Btd RNAi knockdown driven by insc-GAL4 eliminates mature INPs in a subset of type II NB lineages. (C–C′) RNAi knockdown of Pros results in overproliferation of mature INPs. (D–D′) Simultaneous knockdown of Btd and Pros leads to a similar overproliferation of mature INPs in type II NB lineages as Pros RNAi knockdown alone. (E–H′) The loss of mature INPs in btd mutant clones is rescued in pros17 heterozygous mutant larvae. (E–E′) A wild-type type II NB clone has multiple mature INPs. (F–F′) A btd mutant type II NB clone contains no mature INPs. (G–G′) pros17 heterozygous mutant type II NB clone contains multiple mature INPs as the wild-type type II NB clone (A–A′). (H–H′) A btd mutant type II NB clone generated in pros17 heterozygous mutant larvae has multiple mature INPs. Asterisks indicate NBs and arrowheads point to mature INPs in all images. (I–L) Quantifications of the number of mature INPs (I–K) or the percentage of type II NB lineages with mature INPs (J–L) for the rescue of Btd RNAi knockdown phenotypes by Pros RNAi knockdown (I–J) or the rescue of btd mutant phenotypes in pros17 heterozygous mutant larvae (K–L). The sample size on top of each bar represents the number of lineages (I, K, L) or the number of brain lobes (J). The mean and stdev in (L) are calculated by bootstrapping. **, p < 0.01; NS: not significant.
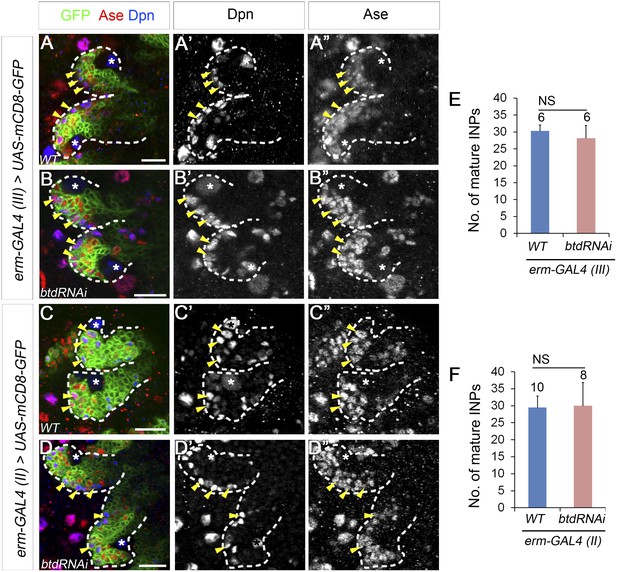
Knockdown of Btd in immature or mature INPs by erm-GAL4 lines does not lead to the loss of mature INPs.
(A–A′′, C–C′′) Wild-type type II NB lineages are labeled with mCD8-GFP driven by erm-GAL4 (III) (A–A′′) or erm-GAL4 (II) (C–C′′). (B–B′′, D–D′′) Knockdown of Btd in Ase+ immature INPs and mature INPs by erm-GAL4 (III) (B–B′′) or in Ase− immature INPs as well as Ase+ immature INP and mature INPs by erm-GAL4 (II) (D–D′′) does not cause a reduction of the number of mature INPs. Only two lineages are shown in each brain. (E–F) Quantifications of the number of mature INPs in type II NB lineages in which Btd is knocked down by erm-GAL4 (III) (E) or erm-GAL4 (II) (F). NS: not significant.
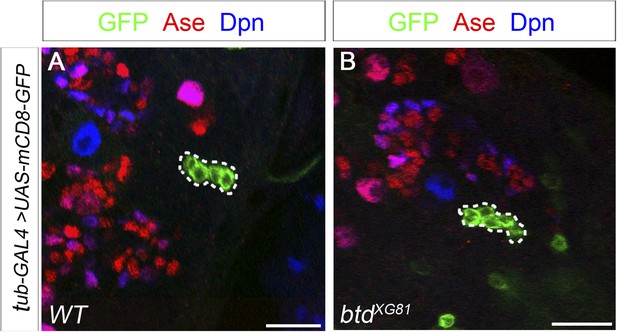
Btd likely does not function in mature INPs.
(A) A wild-type INP clone with 4 post-mitotic cells. (B) A btd mutant INP clone with 6 post-mitotic cells. INP clones labeled with mCD8-GFP are outlined by dashed circles. INP clones are identified as clones that have more than two cells but no NBs.
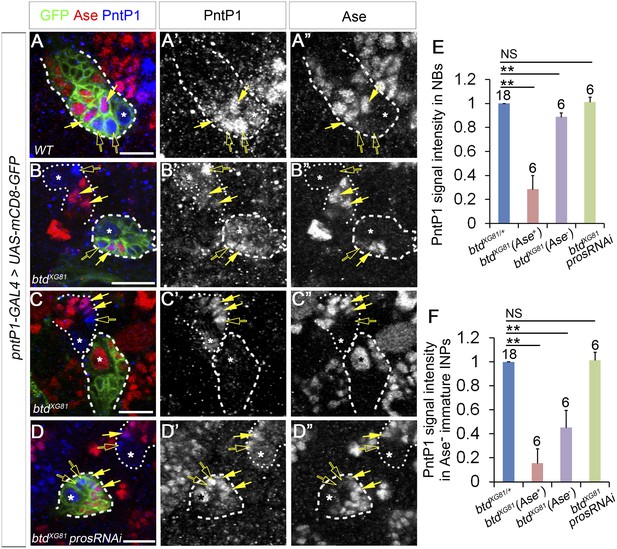
PntP1 expression is reduced in btd mutant type II NB clones.
(A–A′′) PntP1 is expressed in the NB (*), Ase− immature INPs (open arrows), as well as Ase+ immature INPs (solid arrows) in a wild-type type II NB clone. (B–B′′) PntP1 expression is much lower in a btd mutant type II NB clone without the ectopic Ase expression in the NB than that in a neighboring btd heterozygous type II NB lineage. The reduction is particularly obvious in the Ase− immature INPs (open arrows). Note that PntP1 remains expressed in Ase+ daughter cells (arrows) next to the Ase− immature INPs. (C–C′′) PntP1 expression is largely abolished in a btd mutant type II NB clone with the ectopic Ase expression in the NB. In a neighboring btd heterozygous type II NB lineages, PntP1 is still detected in the NBs (*), Ase- immature INPs (open arrows) and Ase+ immature INPs (arrows). (D–D′′) Knocking down Pros restores the expression of PntP1 in the NB (*), Ase− immature INPs (open arrows), and Ase+ immature INPs (arrows) in a btd mutant clone to levels comparable to those in a neighboring btd heterozygous type II NB lineage. Wild-type (A–A′′) or btd mutant type II NB clones (B–D′′) are outlined by dashed lines and neighboring btd heterozygous type II NB lineages (B–D′′) are marked with dotted lines. (E–F) Quantifications of PntP1 expression levels in type II NBs (E) and Ase− immature INPs (F) in btd mutant type II NB clones relative to neighboring type II NB lineages in the same brains. **p < 0.01; NS: not significant.
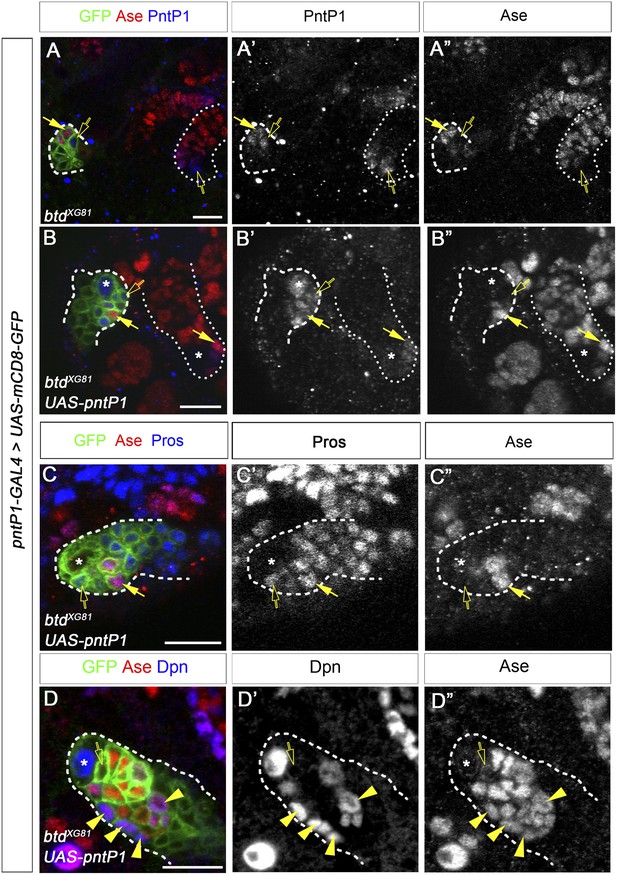
The reduction of PntP1 is unlikely responsible for the loss of INPs in btd mutant type II NB clones.
(A–A′′) A btd mutant type II NB clone has similar expression levels of PntP1 as a neighboring btd heterozygous mutant type II NB lineage but fails to produce mature INPs. (B–B′′) Expressing UAS-PntP1 does not rescue the loss of mature INPs in a btd mutant clone, even though the PntP1 expression in the btd mutant clone is much higher than that in a neighboring btd heterozygous mutant type II NB lineage. (C–C′′) Nuclear Pros remains ectopically expressed in Ase− immature INPs and Ase+ daughter cells in a btd mutant type II NB clone expressing UAS-PntP1. (D–D′′) Mature INPs are partially rescued by UAS-PntP1 in a btd mutant type II NB clone. In all images, btd mutant clones are outlined by dashed lines and their neighboring btd heterozygous mutant type II NB lineages are marked by dotted lines. Open arrows: Ase− immature INPs; solid arrows: Ase+ immature INPs; arrowheads: mature INPs.
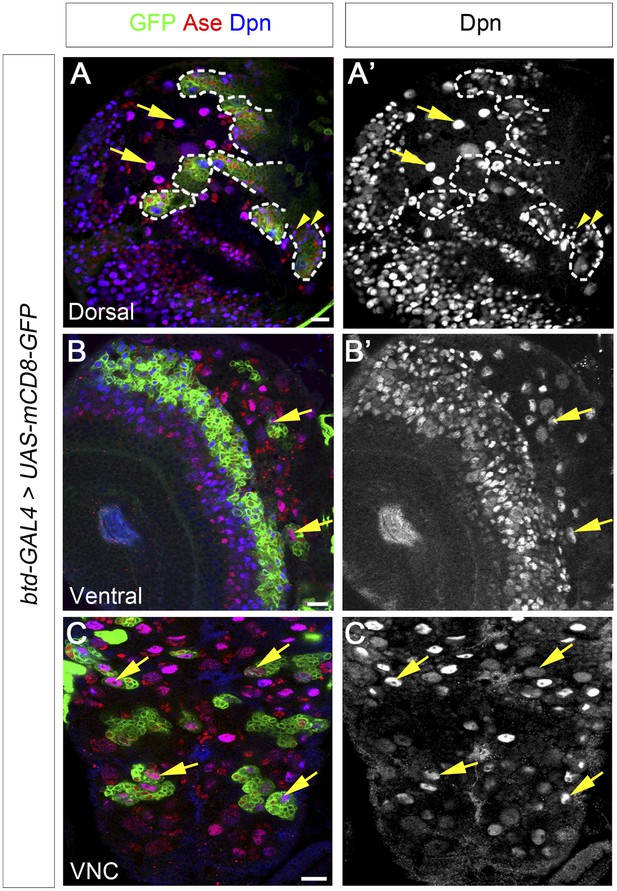
Btd is expressed in type II NB lineages and a subset of type I NB lineages.
(A–A′) mCD8-GFP driven by btd-Gal4 is expressed in all type II NB lineages (outlined by dashed lines) but not type I NB lineages (e.g. arrows) on the dorsal side of a 3rd instar larval brain. The expression of mCD8-GFP becomes progressively weak in cell away from the NB. Some mature INPs (e.g. arrowheads) distant from the NB have no obvious expression of mCD8-GFP. Only seven out of total eight type II NB lineages are shown in this particular focal plane. (B–B′) Two type I NB lineages are labeled by mCD8-GFP driven by btd-GAL4 on the ventral side of a 3rd instar larval brain. (C–C′) mCD8-GFP driven by btd-Gal4 labels a subset of type I NB lineages (e.g. arrows) in the ventral nerve cord (VNC).
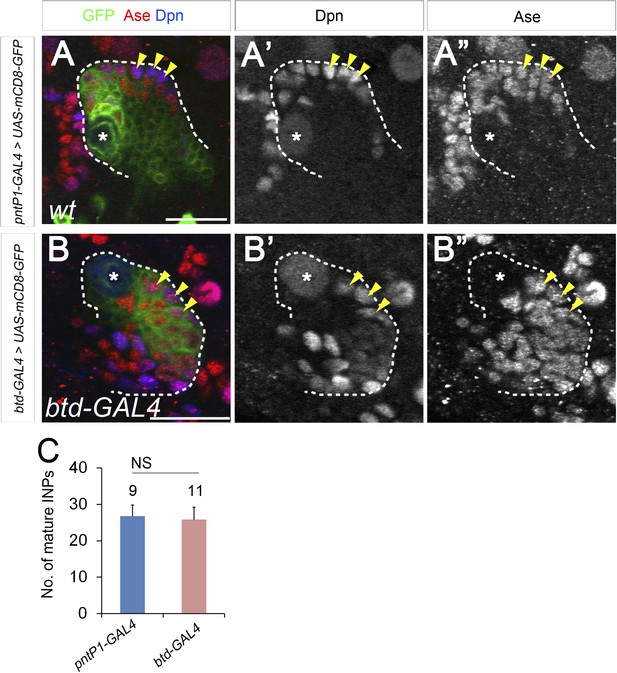
The GAL4 insertion in the btd-GAL4 line does not affect type II NB lineage development.
(A–A′′) A wild-type type II NB clone contains multiple mature INP. (B–B′′) A btd-GAL4 mutant type II NB clone has a similar number of mature INPs as wild-type type II NB clone. (C) Quantifications of the number of mature INPs in wild-type and btd-GAL4 mutant type II NB clones. Arrowheads: mature INPs; NS: not significant.
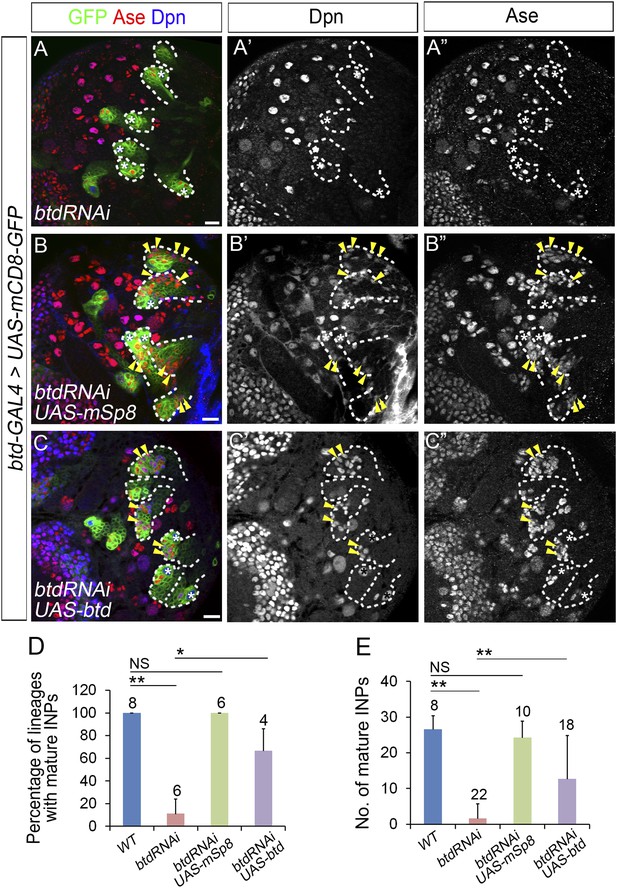
The loss of INP phenotype resulting from Btd RNAi knockdown is rescued by the expression of mouse Sp8 (mSp8) or Drosophila Btd.
(A–A′′) Btd RNAi knockdown driven by btd-GAL4 completely eliminates mature INPs in all type II NB lineages in a 3rd instar larval brain. (B–B′′) The expression of UAS-mSp8 driven by btd-GAL4 fully rescues the loss of mature INPs in all type II NB lineages resulting from Btd RNAi knockdown. (C–C′′) The expression of UAS-btd driven by btd-GAL4 partially rescues the loss of mature INPs in type II NB lineages resulting from Btd RNAi knockdown. Arrowheads point to mature INPs in all images. (D–E) Quantifications of the percentage of type II NB lineages with mature INPs (D) and the number of mature INPs (E) in 3rd instar larvae with indicated genotypes. Sample sizes represent the number of brain lobes (D) or the number of type II NB lineages (E). *, p < 0.05; **, p < 0.01; NS: not significant.
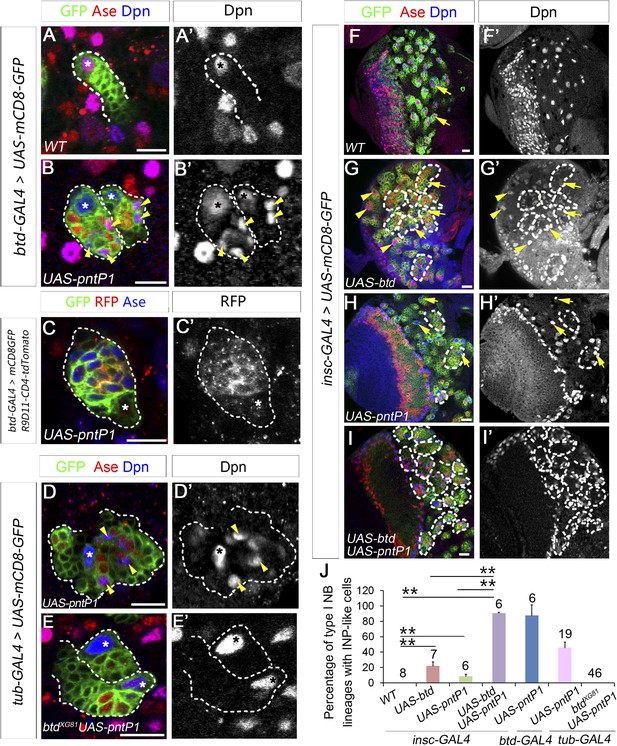
Btd and PntP1 function cooperatively to promote the generation of INPs.
(A–C′) Ectopic expression of PntP1 consistently promotes the generation of INP-like cells in Btd-positive type I NB lineages in larval brains. (A–A′) A wild-type type I NB lineage labeled by btd-GAL4 has no INP-like cells. (B–C′) The ectopic expression of PntP1 driven by btd-GAL4 suppresses Ase in the NB (*) and induces the generation of Ase+ Dpn+ INP-like cells (arrowheads) (B–B′), which also express INP-specific marker R9D11-CD4-tdTomato (C–C′). (D–E′) The expression of PntP1 driven by tub-GAL4 suppresses Ase in both wild-type (D) and btd mutant (E) type I NBs (*) but only induced the generation of INP-like cells in the wild-type type I NB clone (D–D′) but not in the btd mutant clone (E–E′). (F–I′) insc-GAL4 drives the expression of UAS-mCD8-GFP alone (F–F′) or together with UAS-btd (G–G′), UAS-pntP1 (H–H′), or UAS-btd plus UAS-pntP1 (I–I′) in type I NB lineages. Images are from the ventral side of larval brains, where only type I NB lineages are observed in wild-type animals (F–F′). The expression of UAS-btd (G–G′) or UAS-pntP1 (H–H′) alone promotes the generation of INP-like cells only in small subset of type I NB lineages (dashed circles). The expression of Btd only suppresses/reduces Ase expression in type I NBs (arrows) that produce INP-like cells (G–G′) but not in other type I NBs (arrowheads), where PntP1 suppresses Ase in nearly all type I NBs (e.g. arrows) regardless of the generation of INP-like cells (H–H′). Co-expression of Btd and PntP1 promotes the generation of INP-like cells nearly in all type I NB lineages (dashed circles) (I–I′). (J–J′) Quantifications of the percentage of type I NB lineages that produce INP-like cells in larvae with indicated genotypes. The number on top of each bar represents the number of brain lobes except for numbers for the expression of UAS-pntP1 driven by tub-GAL4, which are the number of clones. The mean and stdev for the percentage of wild-type or btd mutant clones expressing UAS-pntP1 driven by tub-GAL4 are calculated by bootstrapping. **p < 0.01.
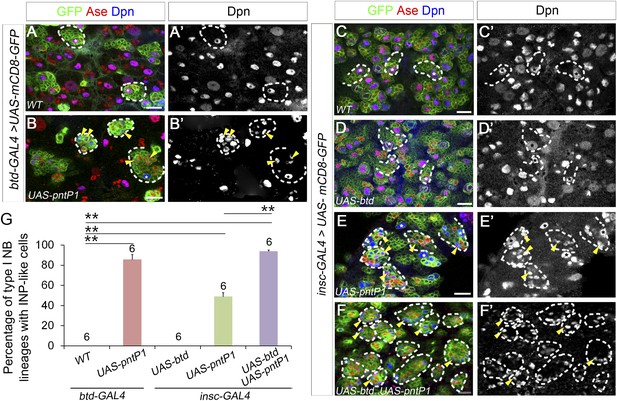
Btd and PntP1 function cooperatively to induce the generation of INP-like cells in type I NB lineages in the VNC.
(A–A′) Type I NB lineages labeled by mCD8-GFP driven by btd-GAL4 in the VNCs have no INPs (e.g. dashed circles). (B–B′) Expression of PntP1 driven by btd-GAL4 induces the generation of INP-like cells (small Dpn+ cells) in nearly all Btd-positive lineages (dashed circles). (C–C′) Type I NB lineages labeled by mCD8-GFP driven by insc-GAL4 in the VNC have no INPs. (D–D′) Expression of UAS-btd driven by insc-GAL4 neither suppresses Ase in the NB nor induces the generation of INP-like cells in type I NB lineages (e.g. dashed circles) in the VNC. (E–E′) Expression of UAS-pntP1 driven by insc-GAL4 suppresses Ase in nearly all NBs (*) and induces the generation of INP-like cells in a subset of type I NB lineages (dashed circles) in the VNC. (F–F′) Co-expression of UAS-btd and UAS-pntP1 driven by insc-GAL4 induces INP-like cells in almost all type I NB lineages (dashed circles) in the VNC. (G) Quantifications of the percentage of type I NB lineages with INP-like cells in brain lobes with indicated genotypes. **p < 0.01.
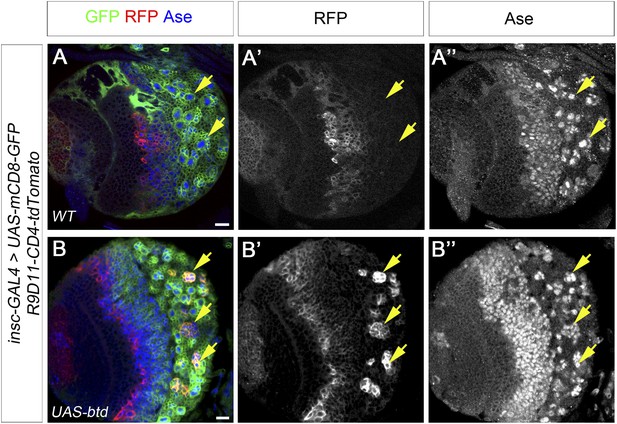
Overexpression of Btd promotes the generation of INP-like cells.
(A–A′′) INP specific marker R9D11-CD4-tdTomato is not expressed in any type I NB lineages (e.g. arrows) on the ventral side of a larval brain. (B–B′′) Overexpression of Btd induces the generation of INP-like cells in a subset of type I NB lineages on the ventral side of a larval brain as indicated by the expression of R9D11-CD4-tdTomato (e.g. arrows).





