A structural mechanism for bacterial autotransporter glycosylation by a dodecameric heptosyltransferase family
Figures
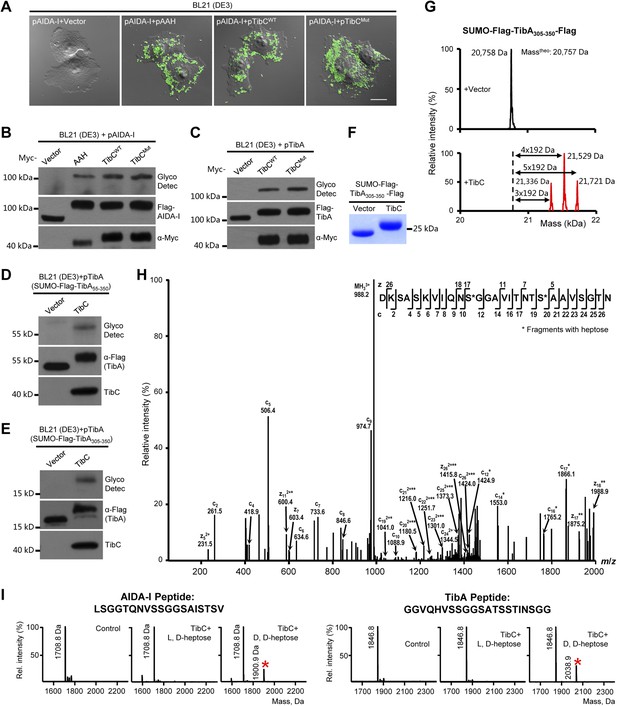
TibC catalyzes TibA/AIDA-I heptosylation and confers AIDA-I-mediated bacterial adhesion to host cells.
(A) E. coli BL21 expressing EGFP was transformed with a plasmid harboring Flag-AIDA-I (pAIDA-I) together with a vector or Myc-tagged autotransporter adhesin heptosyltransferase (AAH)/TibC. TibCWT, wild-type TibC; TibCmut, the TibC mutant used for crystallization. Infected cells and the bacteria were visualized by differential interference contrast microscopy and the green fluorescence, respectively. Scale bar 20 μm. (B and C) Assays of TibC-catalyzed AIDA-I and TibA glycosylation. Lysates of E. coli BL21 (DE3) cells transformed with indicated constructs were subjected to anti-Flag and -Myc immunoblotting or glycosylation analysis. The Flag tag in AIDA-I/TibA was inserted between the signal peptide and the passenger domain. (D–F) Indicated TibA truncations fused C-terminal to the SUMO-Flag were co-expressed with a vector or Myc-TibC in E. coli BL21. Shown in (F) is Coomassie brilliant blue staining of purified SUMO-Flag-TibA305-350-Flag. (G) ESI mass spectrometry analyses of TibC modification of SUMO-Flag-TibA305-350-Flag in the E. coli system. The red peaks mark the glycosylated species. (H) ETD tandem mass spectrum of a triply charged Asp-N digested peptide from TibC-modified SUMO-Flag-TibA305-350-Flag. The fragmentation patterns generating the observed c and z ions are illustrated along the peptide sequence on top of the spectrum. Asterisk marks a modification on the serine by a heptose. (I) In vitro heptosyltransferase activity of TibC. Two peptides derived from the passenger domains of AIDA-I (left) and TibA (right), respectively, were left untreated (control) or reacted with TibC in the presence of indicated sugar ligands. Shown are MALDI-TOF mass spectra of the reacted peptides (*, peptides modified by one heptose).
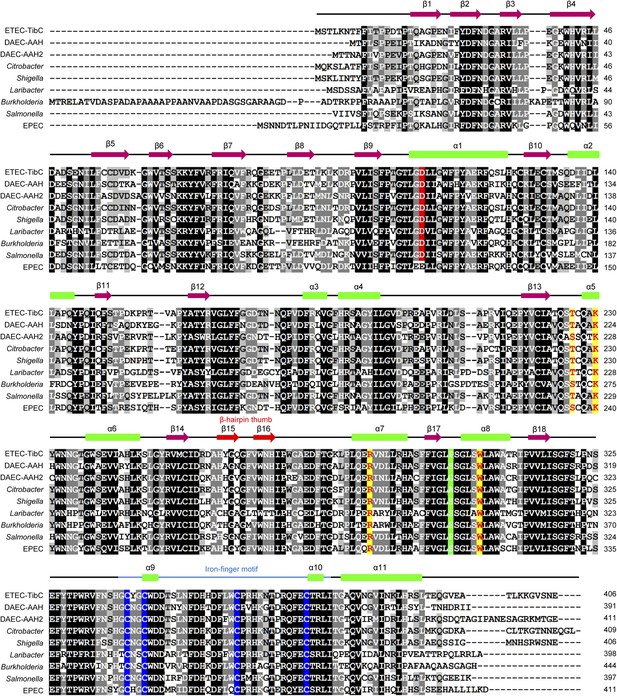
Multiple sequence alignment of the BAHT Family.
Alignment was generated in the GeneDoc program. The genus names are indicated on the left. Secondary structures determined from the TibC crystal are on top of the sequence. The β-hairpin thumb and the iron-finger motif are highlighted as red arrows and a blue line, respectively. Identical residues are in black and other conserved residues are in grey. The ligand binding residues are in yellow and the four iron-finger cysteine residues are in blue. Green color highlights the residue that determines the sugar substrate stereoselectivity, and the catalytic base is in red.

Amino acid sequence of AIDA-I passenger domain (residues 58–588) arranged by β-helix repeat units.
Each row lists the sequence of one repeat unit, which is predicted to form a β-helix composed of three small β-strands (green arrows). The predicated secondary structure is marked on top of the sequence. The solid line represents putative loops. The numbers in each row mark the positions of the amino acids in each repeat unit. Putative glycosylated serine residues are yellow shaded.
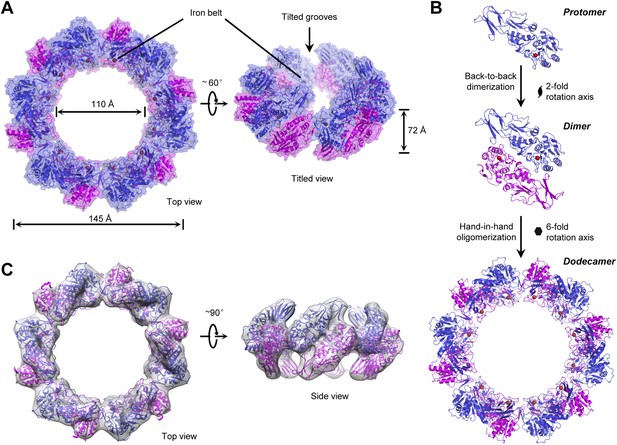
The overall structure and assembly of the TibC dodecamer.
(A) The surface view of the TibC dodecamer. The two adjacent symmetric protomers are colored blue and magenta, respectively. The ferric ions are shown as red spheres. Shown on the left is a top view along the sixfold axis and on the right is a titled view. (B) Assembly of the TibC dodecamer. Two adjacent protomers form a back-to-back dimer with a twofold symmetry. Six symmetric dimers are further assembled through hand-in-hand contacts into a large ring. (C) Cryo-EM reconstruction of TibC at 11.5 Å resolution. The crystal structure was fit into the cryo-EM envelope. Shown are top and side views of the EM structure.
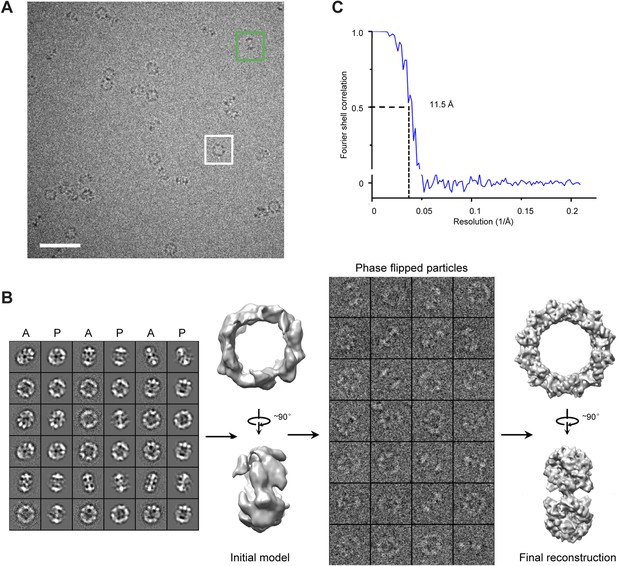
Cryo-EM images and 3D reconstruction of the TibC dodecamer.
(A) The cryo-EM image of TibC dodecamer acquired at a nominal magnification of 75,000×. The white and green boxes indicate typical top and side view particles, respectively. Scale bar, 50 nm. (B) Initial model reconstruction from class averages generated from reference-free classification. C1 symmetry was applied in the reconstruction process. New density was obtained by projection-matching refinement of the starting density using the Fourier reconstruction approach against the phase-flipped particles iteratively with gradually reduced step angle. D6 symmetry was enforced during the refinement. A, average; P, projection. (C) Resolution assessment by Fourier shell correlation (FSC). The FSC coefficient from the final round of the refinement is plotted as a function of spatial frequency (1/Å). The resolution of the final reconstruction is 11.5 Å according to the FSC = 0.5 criterion.
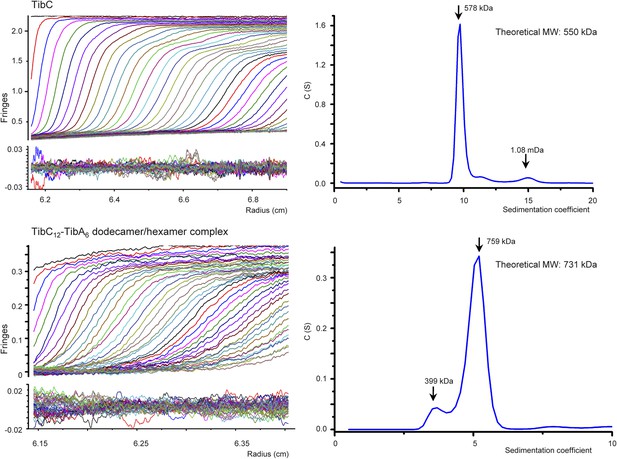
Analytical ultracentrifugation sedimentation velocity analysis of TibC dodecamer and TibC–TibA dodecamer/hexamer complex.
In the left upper panels, fringes were collected by the interference optics and raw sedimentation velocity scans were overlaid with the best-fit curves obtained from sedimentation coefficient distribution analysis. Two or three scans are included and the calculated systematic signal offsets are removed. The left lower panels show the residual fringes after fitting the raw data with modeled curves.
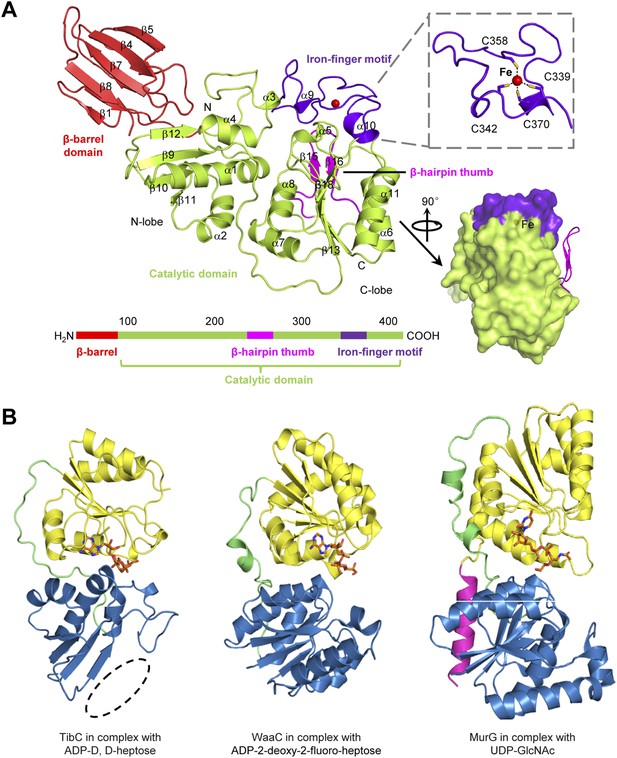
The TibC protomer structure and the glycotransferase domain.
(A) Ribbon diagram of the TibC protomer structure. The upper right shows tetrahedral coordination of the ferric ion by four cysteine residues. Shown on the lower right is a lateral view with the β-hairpin thumb highlighted as magenta ribbons. The domain organization along the primary structure is on the lower left. (B) Structural comparison of TibC with GT-B glycosyltransferases. Left, the catalytic domain of ADP-D,D-heptose-bound TibC structure determined in this study; Center, WaaC bound with ADP-2-deoxy-2-fluoro-heptose (PDB ID: 2H1H); Right, MurG bound with UDP-GlcNAc (PDB ID: 1NLM). The overall structures and sugar ligands are in ribbon diagram and orange sticks, respectively. The N- and C-lobe are colored blue and yellow, respectively. The β-hairpin thumb insertion and the iron-finger motif in TibC are omitted for clarity. The linker between the two lobes and an extra C-terminal helix in MurG are in green and magenta, respectively. The dashed circle highlights the absence of α-helices at the N terminus of the N-lobe in TibC.
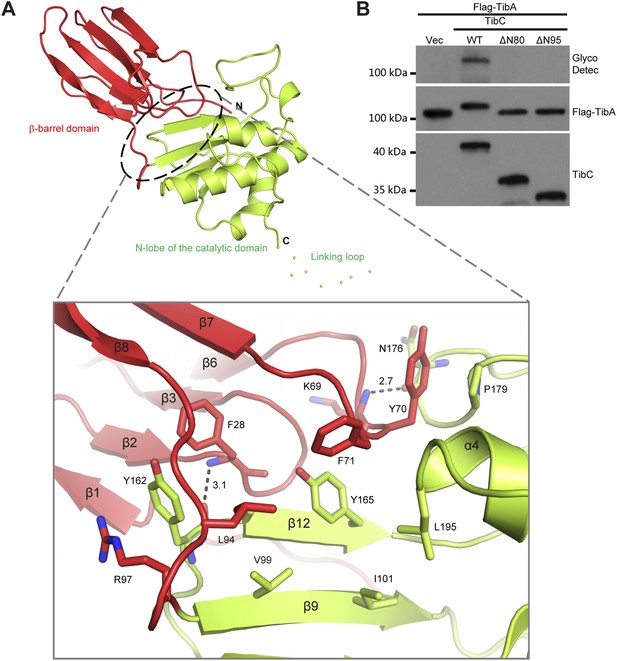
Interaction between the β-barrel and the N-lobe of the catalytic domain in TibC protomer.
(A) Structural interactions between the β-barrel and N-lobe. Shown in the upper panel is the overall structure using the same color scheme as that in Figure 2A. The interface between the two domains is circled with dashed lines and the detailed interactions are in the lower panel with interacting residues in sticks. Polar interactions are represented by black dashed lines with a number denoting the distance in angstrom. R97 and Y162 form a pair-wise cation-π interaction; Y70 and P179 have a π-stacking interaction. Interface residues including F28, F71, L195, I101, V99, L94, and Y165 are clustered together to form a buried hydrophobic core. (B) Effects of β-barrel deletion on TibA glycosylation. TibC or indicated TibC truncations were co-expressed with Flag-TibA in E. coli BL21 (DE3). Expression of TibA and TibC was detected by anti-Flag and anti-TibC immunoblotting, respectively. TibA glycosylation was detected with the ECL glycoprotein detection kit. ΔN80 and ΔN95 refer to deletion of the N-terminal 80 and 95 residues in TibC, respectively.
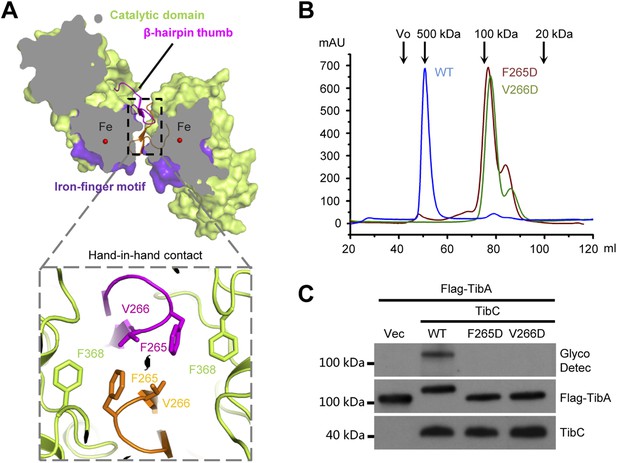
The β-hairpin thumb-mediated hand-in-hand contact required for TibC dodecamer assembly and catalytic activity.
(A) β-hairpin thumb-mediated hand-in-hand contact. Upper, surface presentation of two adjacent TibC protomers with the β-hairpin thumb in ribbons. Lower, interface details with key residues in sticks. Black lens, the twofold axis. (B and C) Effects of hand-in-hand contact mutations on TibC dodecamer formation and catalyzing TibA glycosylation. TibC (WT or indicated mutant) proteins were loaded onto a gel filtration column in (B). Black arrows mark the molecular weight calibration. Vo, void volume. The experiments in (C) were performed and data are presented similarly to those in Figure 1B.
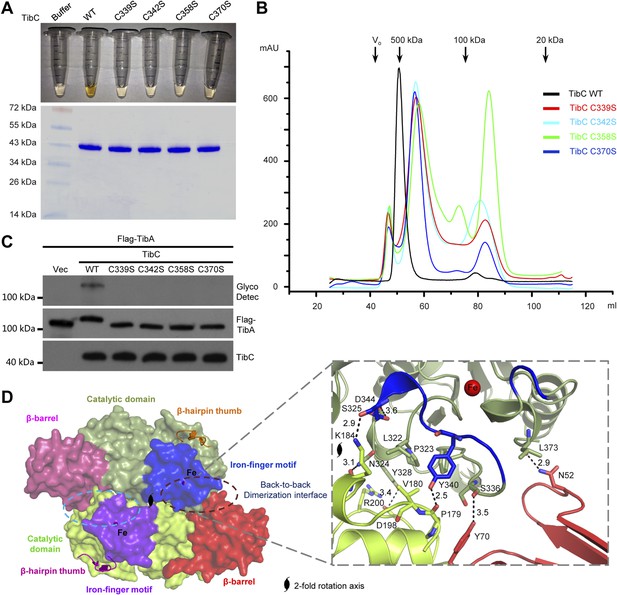
The iron-finger motif critical for TibC dodecamer assembly and heptosyltransferase activity.
(A) Purified TibC (WT and the four iron-finger cysteine mutants, ∼8 mg/ml) were loaded onto a SDS-PAGE gel followed by Coomassie blue staining. (B) Gel filtration chromatography of the iron-finger cysteine mutants of TibC. Black arrows mark the molecular weight calibration and Vo denotes the void volume. (C) Analyses of TibA glycosylation by the iron-finger cysteine mutants of TibC. Indicated proteins were co-expressed in E. coli BL21 (DE3) and the lysates were analyzed by anti-Flag (TibA) and anti-TibC immunoblotting and also TibA heptosylation assay. (D) The back-to-back dimer formation. Shown on the left is the surface presentation colored as indicated. Two symmetric dimerization interfaces are marked by dashed circles. Structural details of one interface are shown on the right with interacting residues in sticks. Polar interactions are represented by black dashed lines with a number denoting the distance in angstrom.
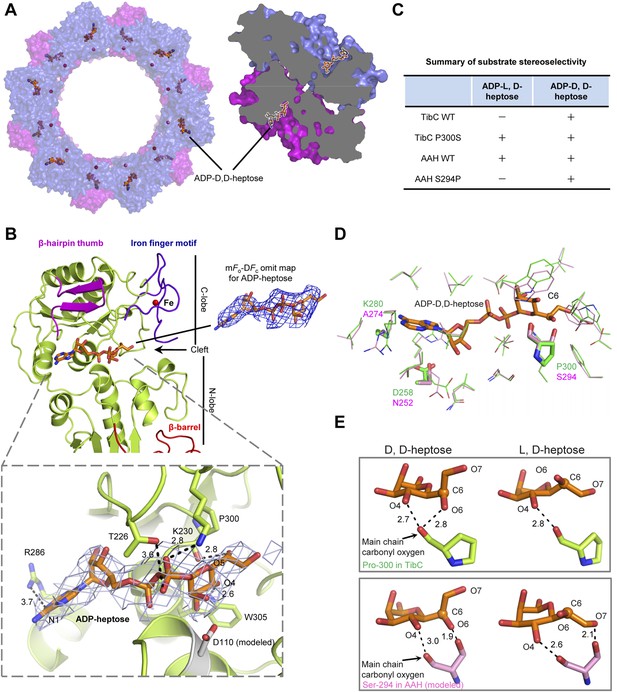
Crystal structure of the TibC-ADP-D,D-heptose complex and sugar ligand stereoselectivity.
(A) Structure of the TibC-ADP-D,D-heptose complex. Left, a top view of the dodecamer. Right, a half-cut view of the back-to-back dimer. ADP-D,D-heptose and ferric ions are shown as ball-and-stick models and red spheres, respectively. (B) Binding of ADP-D,D-heptose to TibC protomer. Key sugar-binding residues are in sticks. Polar interactions are indicated by black dashed lines with a number denoting the distance in angstrom. ADP-D,D-heptose is shown as orange sticks meshed with σA-weighted 2mFo-DFc electron density map contoured at 1.0 σ. The mFo–DFc omit density of the ligand at a contour level of 3.0 σ is shown in blue on the right. (C) Sugar ligand stereoselectivity and effects of swapping Pro-300 in TibC with the corresponding Ser-294 in autotransporter adhesin heptosyltransferase (AAH). The raw experimental data are in Figure 6—figure supplement 2A. (D) Comparison of ADP-D,D-heptose binding residues in TibC (green) and AAH (pink). The AAH structure was modeled from that of TibC. Conserved residues are shown as lines; divergent residues are in sticks. ADP-D,D-heptose is in orange sticks with the chiral C6 as a sphere. (E) A proposed model for the ligand stereoselectivity. The stereoselectivity determinants (Pro-300 in TibC and Ser-294 in AAH) are in sticks. The distance between the sugar ligand and the Pro-300/Ser-294 are denoted as a number in angstrom.
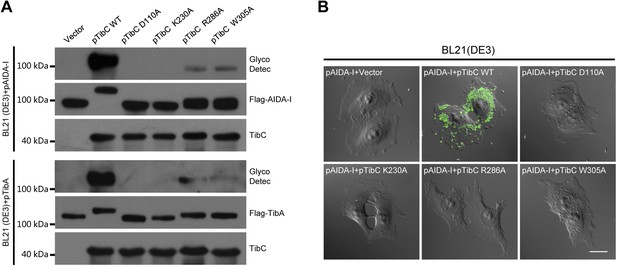
Requirement of TibC heptosyltransferase activity for bacterial adhesion to host cells.
(A) Assays of TibA and AIDA-I glycosylation by TibC mutants in E. coli. BL21 (DE3) cells were transformed with a Flag-TibA (upper) or AIDA-I (lower) expression plasmid alone (vector) or together with an indicated TibC mutant construct (pTibC). TibA and AIDA-I glycosylation was detected with the Amersham ECL glycoprotein detection kit. (B) Assays of TibC mutants in supporting bacterial adhesion to HeLa cells. BL21 (DE3) cells were transformed with an AIDA-I expression plasmid together with an indicated TibC mutant. Adhesion assay was performed and data were presented similarly to those in Figure 1A. Scale bar, 20 μm.
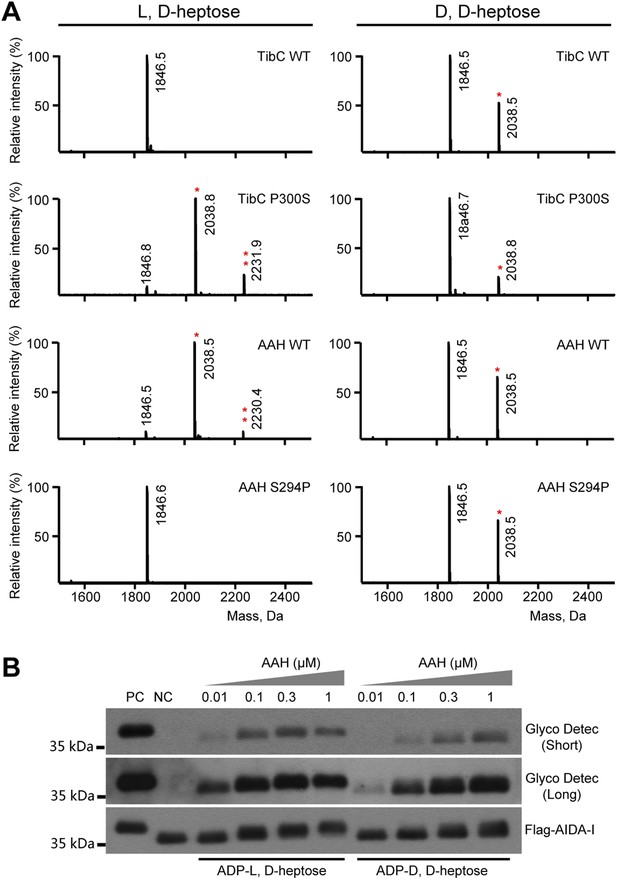
Assays of the ligand stereoselectivity of TibC and autotransporter adhesion heptosyltransferase (AAH).
(A) A TibA-derived synthetic peptide (GGVQHVSSGGSATSSTINSGG) (theoretical molecular weight 1846.5 Da) was reacted with wild-type (WT) or indicated TibC or AAH mutants in the presence of indicated sugar donors. The peptides were then subjected to MALDI-TOF mass spectrometry analysis and shown are the spectra obtained. Asterisks mark the peptides modified by one (*) or two (**) heptose molecules. (B) In vitro heptosylation assays of recombination AAH using GST-AIDA-I531-611-Flag substrate. 1 μM GST-AIDA-I531-611-Flag, 100 μM indicated sugar ligand and recombinant AAH with a concentration gradient (from 0.01 μM to 1 μM) were used. The reaction mixtures were subjected to immunoblotting and glycoprotein detection analyses. Purified GST-AIDA-I531-611-Flag and GST-AIDA-I531-611-Flag coexpressed with AAH were used as negative (NC) and positive control (PC). Short and long exposures of the glycosylated AIDA-I are shown.
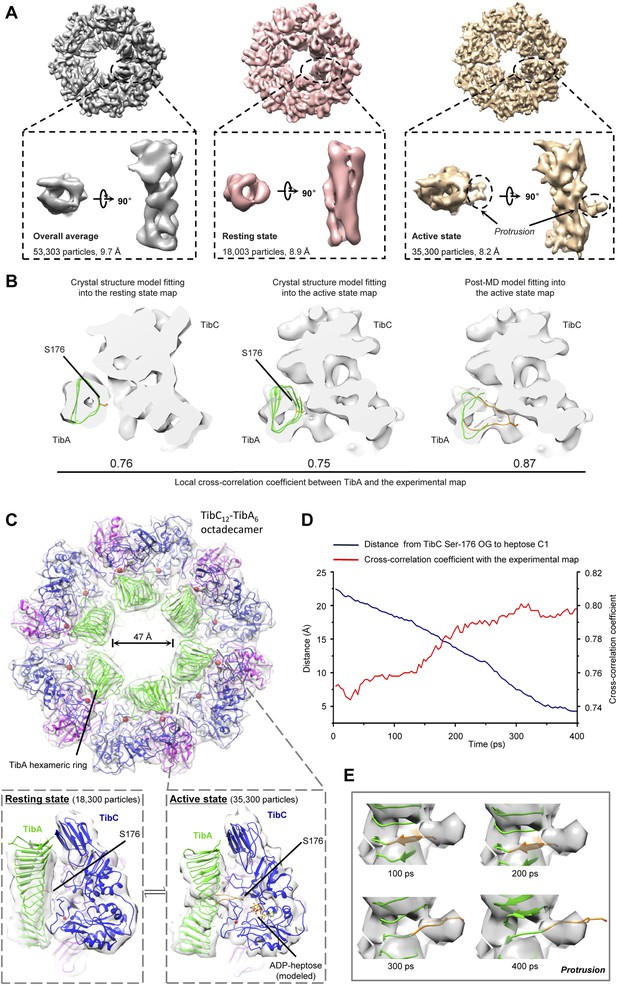
Cryo-EM models of the TibC–TibA55-350 dodecamer/hexamer complex.
(A) Cryo-EM maps of the TibC–TibA55-350 complex. The left is the overall average map obtained from classification-free reconstruction. A two-class classification was employed to distinguish different features observed with the TibA density (Figure 7—figure supplement 2), and the resulting two maps are shown in the middle and right panels. (B) Model fitting of TibA55-350 crystal structure into the two classes of experimental maps and comparison with the post-molecular dynamics (MD) model. The cross-correlation coefficient (CCC) between TibA and the experimental map is listed underneath the models. The protrusion loop is highlighted in orange and the to-be-glycosylated Ser-176 is in sticks. (C) Cryo-EM reconstruction of the TibC–TibA dodecamer/hexamer complex. The atomic model after MD simulation was fit into the cryo-EM map (the upper panel). Ribbon diagram of TibC dodecamer is colored as that in Figure 2A. The lower panel shows the TibA–TibC2 trimer in the active and resting states. ADP-heptose and Ser-176 are in sticks. The conformationally changed TibA loop is highlighted in orange. (D) The CCC for TibA during the steered MD simulation. The distance from TibA Ser-176 OG to heptose C1 (blue line) and the local CCC value for TibA (red line) are plotted as a function of time. (E) Protrusion of the TibA loop sampled by the steered MD simulation. The snapshots at 100, 200, 300, and 400 ps were fitted into the active state cryo-EM map. The protrusion loop is in orange and Ser-176 is in sticks.
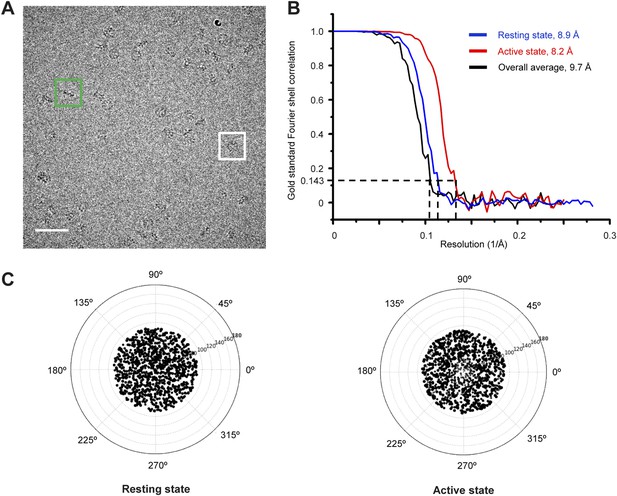
Cryo-EM images of the TibC-TibA55-350 complex.
(A) The cryo-EM image of TibC–TibA55-350 complex acquired at a nominal magnification of 75,000×. The white and green boxes indicate typical top and side view particles, respectively. Scale bar, 50 nm. (B) Fourier shell correlation (FSC) of cryo-EM reconstruction of the overall, resting, and active conformation states. Using the gold standard FSC = 0.143 criterion, the resolutions for the overall average, resting, and active states are 9.7 Å, 8.7 Å, and 8.2 Å, respectively. (C) Euler angle distribution plot of the resting and active conformation states. The size of the black dot is proportional to the number of particles that belong to the specific Euler angle.
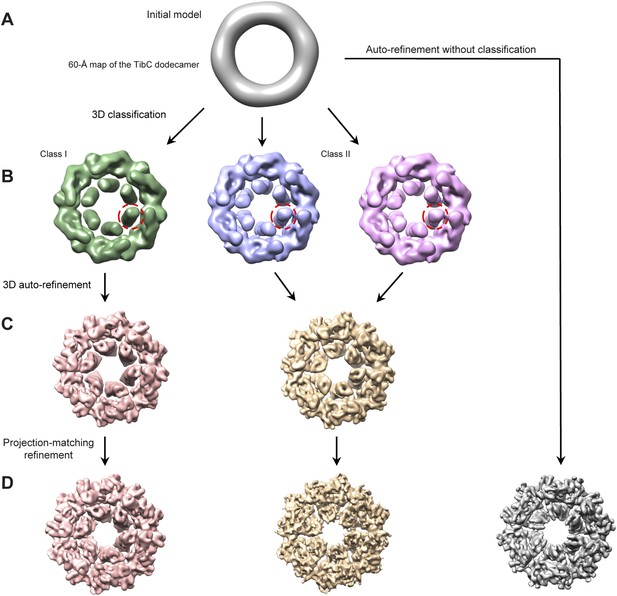
Cryo-EM reconstruction of the TibC12-TibA6 complex.
(A) Low-pass filtered TibC dodecamer was used to initiate the reconstruction. (B) After 3D classification, three conformational states appeared. Two similar ones (middle and right) were merged into one super class (Class II) due to their similar TibA density (red circles). The two classes were then subjected to independent auto-refinement. (C) Following the auto-refinement, the two reconstructed density maps showed more evident discrepancy. Particles in each class were sorted into two independent stacks and subjected to further projection-matching refinement. (D) The resulting final maps had higher resolution with more structural details compared with the overall average map (gray) obtained by auto-refinement and reconstruction without classification.
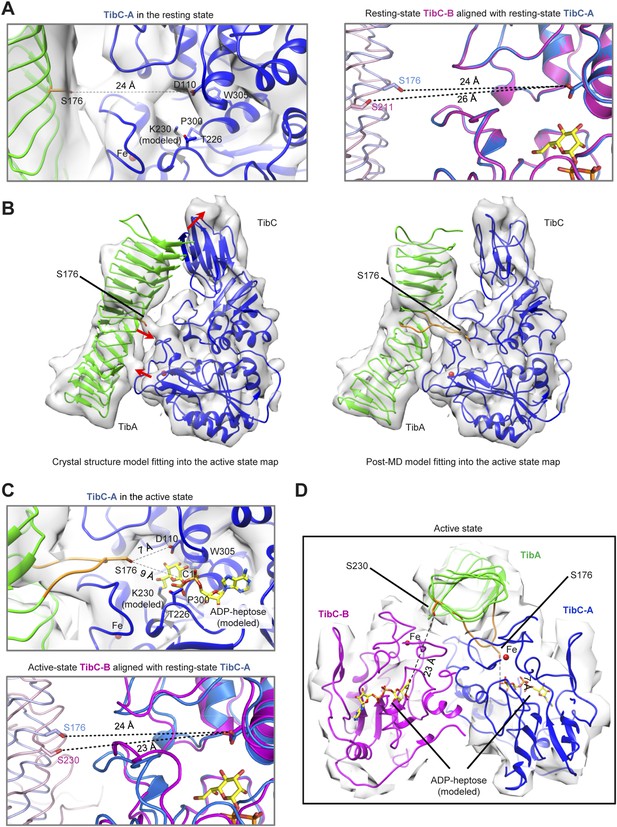
Molecular dynamics (MD)-assisted cryo-EM analysis reveals two conformational states of the TibC–TibA complex.
(A) Catalytic center of the resting state TibC–TibA55-350 complex. Left, structure of one TibC protomer (TibC-A) in a back-to-back dimer. Right, structure of the other TibC protomer (TibC-B) aligned with TibC-A using the Cα trace as the reference. The dashed line with a number in angstrom marks the candidate substrate serine to Asp-110 or heptose C1. (B) Comparison of the rigid body crystal structure model fitting and the post-MD model fitting into the active state EM map. Red arrows mark the major structural differences. (C) Catalytic center of the active state TibC–TibA55-350 complex. Upper, the active state TibC protomer (TibC-A) in a back-to-back dimer. Lower, structure of the other TibC protomer (TibC-B) aligned with the resting state TibC-A using the Cα trace as the reference. The dashed line with a number in angstrom marks the candidate substrate serine to Asp-110 or heptose C1. (D) Overall structure of one TibC back-to-back dimer (blue and magenta) bound with one TibA (green) in the active state.
Tables
Data collection and refinement statistics
| Crystals | TibC-SeMet | TibC D110A in complex with D,D-heptose |
|---|---|---|
| Data collection | ||
| Space group | P21 | P21 |
| Wavelength (Å) | 0.9789 | 0.9792 |
| a, b, c (Å) | 87.8, 314.4, 164.5 | 83.3, 313.2, 164.7 |
| α, β, γ (º) | 90, 101.4, 90 | 90, 101.3, 90 |
| Resolution range (Å)* | 20.0–2.90 (2.95–2.90) | 20.0–3.87 (3.94–3.87) |
| No. of unique reflections | 194,303 (9760) | 79,888 (3968) |
| Completeness (%) | 99.9 (99.9) | 99.9 (100) |
| Redundancy | 7.3 (6.1) | 4.6 (4.6) |
| I/σI | 19.5 (2.6) | 9.18 (2.0) |
| Rmerge (%) | 12.7 (98.6) | 23.7 (92.8) |
| Refinement statistics | ||
| Rwork/Rfree (%)† | 20.8/24.4 | 26.4/27.6 |
| No. of protein atoms | 37,201 | 37,136 |
| No. of ligands atoms | 32 | 504 |
| No. of waters | 5 | 0 |
| RMSD bond lengths (Å) | 0.013 | 0.006 |
| RMSD bond angles (º) | 1.40 | 1.11 |
| Average overall B-factor | 69.10 | 150 |
| Iron atoms B-factor | 58.90 | 150 |
| ADP-heptose B-factor | 150 | |
| Ramachandran plot statistics | ||
| Most favored regions (%) | 94.51 | 94.73 |
| Additional allowed regions (%) | 5.36 | 4.45 |
| Outlier regions (%) | 0.13 | 0.83 |
-
*
The data for the highest resolution shell are shown in parentheses.
-
†
Rfree is calculated by omitting 5% of the total number of reflections in model refinement.
-
RMSD, root-mean-square deviation; SeMet, selenomethionine;





