Transposition-mediated DNA re-replication in maize
Figures
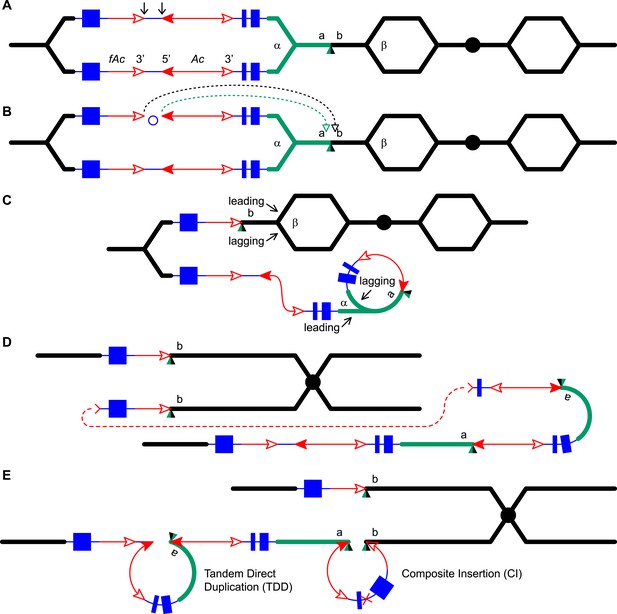
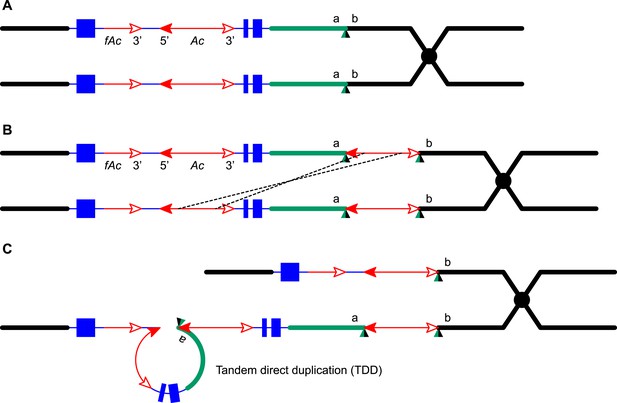
Reversed Ac ends transposition (RET) during DNA replication generates Tandem Direct Duplication (TDD) and Composite Insertion (CI).
Lines indicate a replicating chromosome, hexagons indicate replicons. The blue boxes are exons 1, 2, and 3 (right to left) of the p1 gene, and the green/black triangles are the transposition target site. Red lines with arrow(s) indicate Ac/fAc insertions, and the open and solid arrowheads indicate Ac/fAc 3′ and 5′ ends, respectively. Two replication forks considered here are marked α and β. For animated version, see Video 1. (A) The locus containing fAc/Ac is replicated. Vertical arrows indicate the sites of Ac transposase cuts at the fAc 3′ and Ac 5′ ends. (B) Transposase cleaves and the inter-transposon segment is ligated to form a circle. The excised transposon ends will insert into an unreplicated target site indicated as the green/black triangle. Like standard Ac/Ds transposition, insertion of the Ac/fAc termini into the target site generates an 8-bp target site duplication (TSD; green/black triangle). (C) Insertion of the excised transposon termini places fAc and fAc-flanking DNA ahead of replication fork β (upper chromatid), and Ac and Ac-flanking DNA ahead of replication fork α to generate a rolling circle replicon (lower chromatid). DNA replication continues. (D) Following re-replication of fAc, Ac, and a portion of the flanking sequences, DNA replication forks α and β stall and abort, resulting in chromatids terminated by broken ends (the red > or < symbol) (Michel et al., 1997). The dotted red line connects the two broken ends that will fuse together. (E) Chromatid fusion produces a chromosome with two unequal sister chromatids: The upper chromatid contains a deletion of the segment from fAc to the a/b target site. The lower chromatid contains a TDD (left-hand loop), as well as a new CI (right-hand loop). The TDD contains the DNA deleted from the upper chromatid; the CI contains the re-replicated Ac, fAc and flanking sequences. The junction where broken chromatid ends were joined is indicated by the red ×.
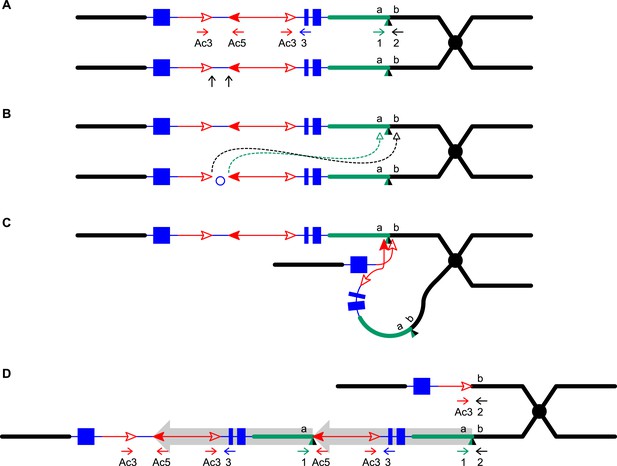
Reversed Ac ends transposition after DNA replication generates Tandem Direct Duplications (TDDs).
The two lines indicate sister chromatids of fully replicated maize chromosome 1, joined at the centromere (black). The blue boxes are exons 3, 2, and 1 (left to right) of the p1 gene. Red lines with arrowhead(s) indicate Ac/fAc insertions, and the open and solid arrowheads indicate the 3′ and 5′ ends, respectively, of Ac/fAc. The short horizontal arrows show the orientations and approximate positions of PCR primers, and the numbers below are the primer names. The green/black triangles indicate the transposon target site sequences and target site duplications. (A) Ac transposase cleaves the lower chromatid at the 3′ end of fAc and the 5′ end of Ac (arrows). (B) Following transposase cleavage, the internal p1 genomic sequences are joined to form a circle. Dotted lines indicate the insertion of the fAc and Ac termini into the a/b site on the sister chromatid. (C) Transposon ends insert into the upper sister chromatid at the a/b target site. (D) The Ac 5′ end joins to the distal side (green) of the target site and the fAc 3′ end joins to the proximal side (black) of the target site to generate a proximal deletion (upper chromatid) and a direct duplication (lower chromatid). The shaded arrows encompass the duplicated segments. Note: this Figure is adopted from Figure 1 of Zhang et al. (2013).
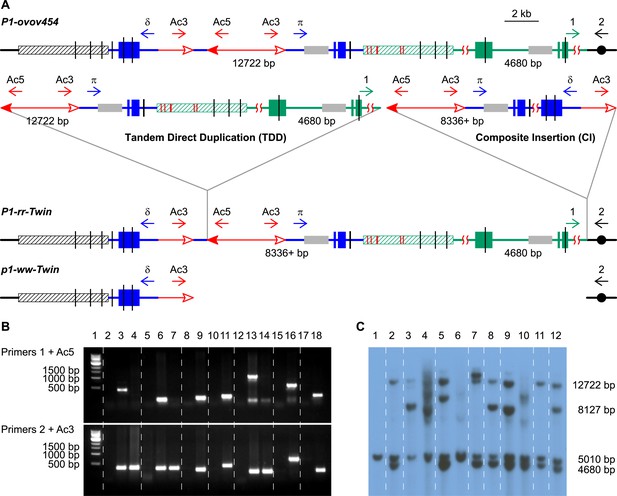
PCR screening and DNA gel blotting of candidate TDDCI alleles.
(A) Detailed structures of P1-ovov454 (progenitor) and RET-generated P1-rr-twin/p1-ww-twin (TDDCI/Deletion) alleles deduced from Figure 1. The horizontal blue lines are p1 gene sequence while the green lines are p1 proximal sequences, including the p2 gene sequence (a p1 paralog, ∼70 kb proximal to p1); the blue and green boxes are exons 1, 2, and 3 (right to left) of p1 and p2, respectively. The small horizontal arrows indicate the orientation and the approximate position of the PCR primers. The gray boxes indicate probe 8B used in DNA gel blot analysis, the short vertical black lines are SacI sites, and the numbers between the SacI sites indicate the lengths of those fragments detected by probe 8B. The hatched boxes represent the distal (black) and proximal (green) 5248 bp repeats flanking the p1 locus. These repeats are identical except for six SNPs, indicated by short red vertical lines inside the green hatched box (SNPs 3 and 4 are only 43 bp apart). Other symbols have the same meaning as in Figure 1. (B) PCR products obtained using primers 1 + Ac5 (upper) or 2 + Ac3 (lower). Lane 1, 1 kb DNA ladder; lane 2, P1-ovov454; lane 3, P1-rr-T22; 4, p1-ww-T22; lane 5, P1-ovov454; lane 6, P1-rr-T24; 7, p1-ww-T24; lane 8, P1-ovov454; lane 9, P1-rr-E17; lane10, P1-ovov454; lane 11, P1-rr-E340; lane 12, P1-ovov454; lane 13, P1-rr-T21; 14, p1-ww-T21; lane 15, P1-ovov454; lane 16, P1-rr-E5; lane 17, P1-ovov454; lane 18, P1-rr-E311. Note: the sequences of primers 1 and 2 are specific for each allele. (C) DNA gel blot analysis of the TDDCI/deletion alleles. Genomic DNA was digested with SacI and the blot was hybridized with probe 8B (see Figure 2A for the position of the probe). Lane 1: p1-ww[4Co63], lane 2: P1-ovov454/p1-ww[4Co63], lane 3: P1-rr-T22/p1-ww[4Co63], lane 4: p1-ww-T22/p1-ww[4Co63], lane 5: P1-rr-T24/p1-ww[4Co63], lane 6: p1-ww-T24/p1-ww[4Co63], lane 7: P1-rr-E17/p1-ww[4Co63], lane 8: P1-rr-E340/p1-ww[4Co63], lane 9: P1-rr-T21/p1-ww[4Co63], lane 10: p1-ww-T21/p1-ww[4Co63], lane 11: P1-rr-E311/p1-ww[4Co63], lane 12: P1-rr-E5/p1-ww[4Co63].

An ear with twinned sectors.
The photo shows two sides of the same ear. Left-side view has a large area with parental P1-ovov454 phenotype (orange pericarp with frequent colorless sectors), while the right-side view shows a large area with typical P1-rr-Twin phenotype (dark red pericarp with few colorless sectors). A single large p1-ww-Twin sector (kernels with mostly colorless pericarp) is visible in both views. The solid purple kernels present in all the sectors result from an independent germinal reversion of the r1-m3::Ds allele and can be ignored.
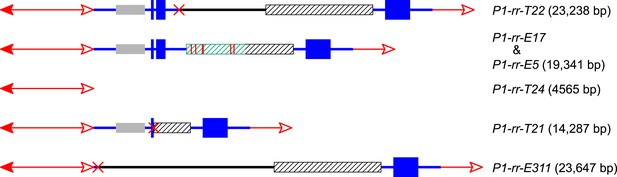
The structures and sizes of Composite Insertions (CIs).
The double-headed arrows (left side) indicate Ac elements, while the single-headed arrows (right side) indicate fAc. The red × symbol indicates the junction of the two re-replicated segments in the insertion. Other symbols have the same meaning as in Figure 1 and Figure 2A.
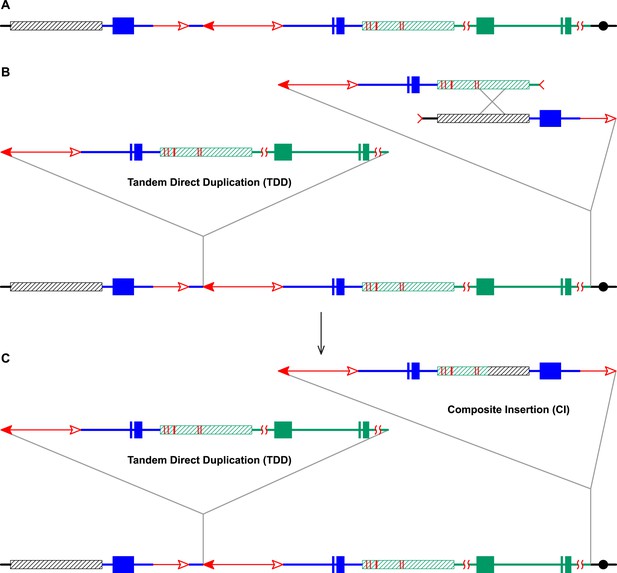
RET followed by homologous recombination generates identical 19,341 bp Composite Insertions in P1-rr-E17 and P1-rr-E5.
(A) Structure of the chromosome 1S segment containing the progenitor P1-ovov454 allele, prior to RET. (B) Drawing shows the RET stage corresponding to Figure 1D. Recombination between the 5248 bp repeats near the two DSBs (marked by > or <) generates a Composite Insertion. (C) Structure of P1-rr-E17 containing TDD (left-hand triangle) and Composite Insertion (right-hand triangle). All the symbols have the same meaning as in Figure 2. Note: P1-rr-E5 contains the 19,341 bp CI but does not contain the TDD. See text for details.
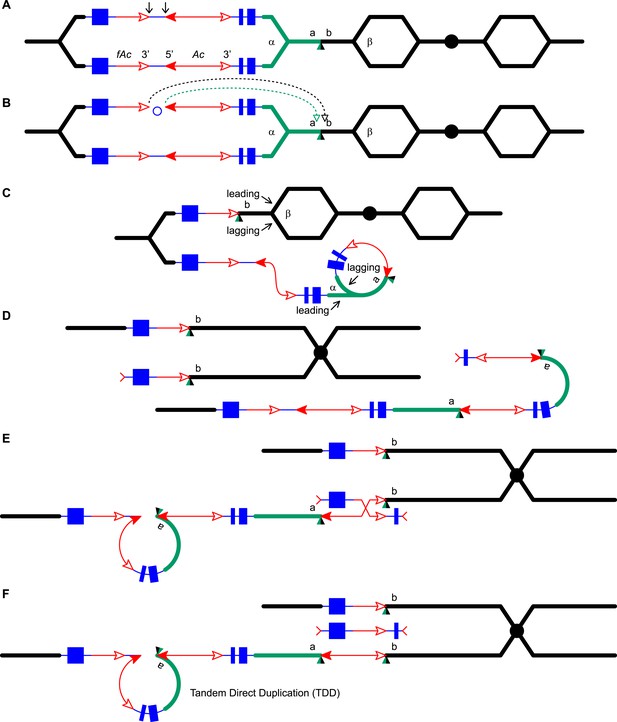
RET followed by homologous recombination generates a simple Ac insertion in P1-rr-T24.
(A), (B), (C), and (D) are the same as in Figure 1. (E) Homologous recombination occurs between the re-replicated Ac and fAc. (F) Two new chromatids are formed: the lower chromatid contains a Tandem Direct Duplication and an Ac insertion, and the upper chromatid carries a reciprocal deletion. For animated version, see Video 2.
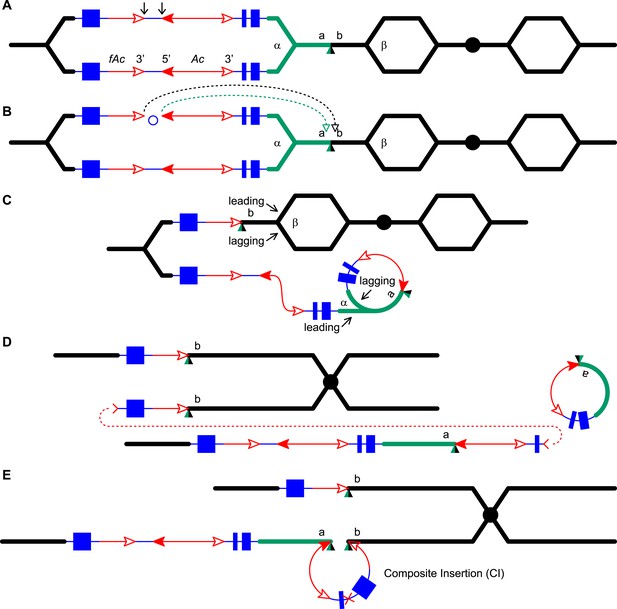
Generation of a Composite Insertion in the absence of a duplication.
(A), (B), and (C) are the same as in Figure 1. (D) Upper chromatid contains deletion; in lower chromatid stalling and abortion of rolling circle replication fork releases the circle. (E) The two chromatids fuse to form a new chromatid containing a Composite Insertion. For animated version, see Video 3.
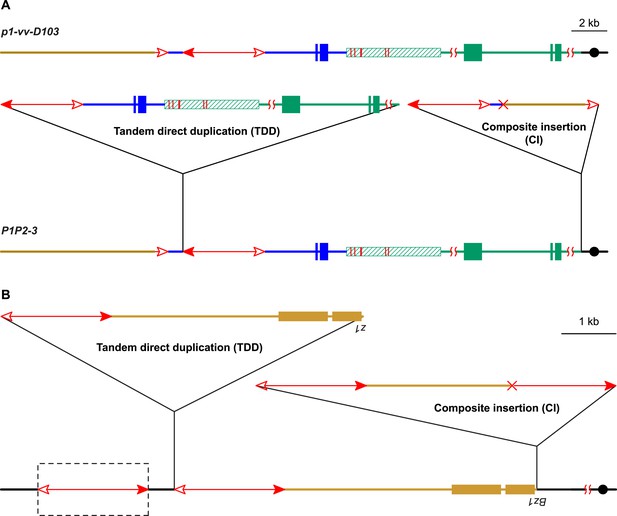
Two additional maize alleles likely generated by RET and re-replication.
(A) Structure of progenitor allele p1-vv-D103 (upper) and TDDCI allele P1P2-3 (lower). The p1-vv-D103 allele is carried on a chromosome 1–10 translocation; the brown line indicates DNA segment from chromosome 10. See text for details. Other symbols have the same meaning as in previous figures. (B) TDDCI structure of bz1-m4-D6856. The bronze-colored boxes indicate exons 1 and 2 (right to left) of the bronze1 gene on maize chromosome 9. The baseline shows the predicted structure of the progenitor of bz1-m4-6856. The dashed box encloses a hypothetical Ds element proposed to have been involved in the generation of bz1-m4-D6856 via RET. For animation, see Video 4. Other symbols as in previous figures. The structure of bz1-m4-D6856 is deduced from Dowe et al. (1990) and Klein et al. (1988).
NAHR generates Tandem Direct Duplications.
All the symbols have the same meanings as in Figure 1. (A) Ac transposes to a site between a and b. (B) Homologous recombination between two non-allelic Ac elements on sister chromatids generates a deletion (upper chromatid) and a TDD (lower chromatid) in (C).
Videos
Animation showing model for reversed Ac ends transposition during DNA replication.
See Figure 1 legend for details.
Animation showing model for RET followed by homologous recombination and generation of a simple Ac insertion in P1-rr-T24.
See text for details.
Animation showing model for RET followed by NHEJ repair and generation of a CI in P1-rr-E311 and P1-rr-T21.
The CI in P1-rr-E5 was generated via a similar mechanism (i.e. the rolling circle was released when forming a DSB), but the DSBs were repaired by homologous recombination as shown in Figure 5 (without the TDD). See text for details.
Animation showing model for generation of TDDCI structure of bz1-m4-D6856 via rolling circle replication.
See Figure 8B legend for details.
Tables
Features of alleles generated by RET-induced DNA re-replication
| Allele number | Allele type | Distance from donor locus to CI* |
|---|---|---|
| P1-rr-T21 | Solo-CI | 13,392 bp |
| P1-rr-E5 | Solo-CI | 16,497 bp |
| P1-rr-T22 | TDDCI | 70 kb |
| P1-rr-T24 | TDDCI | 80 kb |
| P1-rr-E340 | TDDCI | 447 kb |
| P1-rr-E311 | Solo-CI | 563 kb |
| P1-rr-E17 | TDDCI | 1.7 Mb |
-
*
Distance given is from the 5′ end of Ac in the progenitor P1-ovov454 allele, to the point of insertion of the CI; that is, the distance between the TDD and CI insertion points in Figure 2A. In TDDCI alleles, this distance is also the length of the duplicated segment. Except for the fully sequenced alleles P1-rr-T21 and P1-rr-E5, the values given are based on the B73 reference genome sequence (Schnable et al., 2009), which likely differs from the genotype used in these experiments.
Primer sequences
| Primer 1 | P1-rr-T22 | CTGTGGTCGTCCTGCTCCG |
| P1-rr-E17 | AGATTTGACAGAACAGCCCGCAC | |
| P1-rr-T24 | GGTCACGCCCATAATAAAACAATAC | |
| P1-rr-E340 | AACCCGTCTCATCATCATCAGTGT | |
| P1-rr-T21 | GGTTTGTTTGTGCTGCCTCC | |
| P1-rr-E311 | TCGTTCTCTGGTTGGTCGTCGT | |
| P1-rr-E5 | ATTGGTCCCTCCCTCTCCCT | |
| Primer 2 | P1-rr-T22 | AGAACTACTGGAACTCGCACCTCA |
| P1-rr-E17 | CCAGAGTATAGGGTCATGGAGCC | |
| P1-rr-T24 | GCGTCCTCTATCCATTCACTTTCA | |
| P1-rr-E340 | TTTATGAGCCGCTGAATCGC | |
| P1-rr-T21 | CCGATGCTCTTTTCCTTCTCTTCC | |
| P1-rr-E311 | GCGATGCTATCAGTTAGACCAGGC | |
| P1-rr-E5 | CGCCGAACTTTCACTGCTCTGCTA | |
| Ac3 | GATTACCGTATTTATCCCGTTCGTTTTC | |
| Ac5 | CCCGTTTCCGTTCCGTTTTCGT | |
Additional files
-
Supplementary file 1
Sequences flanking the Composite Insertion in the TDDCI and solo-CI alleles and sequences flanking fAc in the deletion alleles.
- https://doi.org/10.7554/eLife.03724.019





