Evidence for suppression of immunity as a driver for genomic introgressions and host range expansion in races of Albugo candida, a generalist parasite
Figures
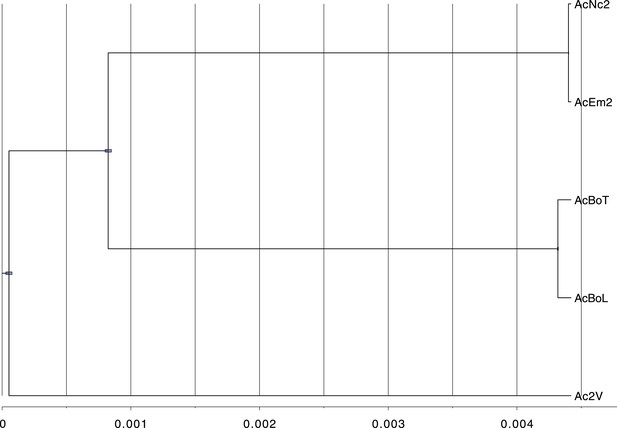
Phylogeny shows that the five sequenced Albugo candida isolates fall into three divergent races.
BEAST tree constructed based on using contig 1 (398,508 bp) shows the divergence among the three A. candida races (AcEm2 and AcNc2; AcBoT and AcBoL; Ac2V). Blue bars represent the 95% Higher Posterior Density (HPD). Here, we used a strict molecular clock fixed at 1.0 in order to show the relationship in the scale of substitutions per site.
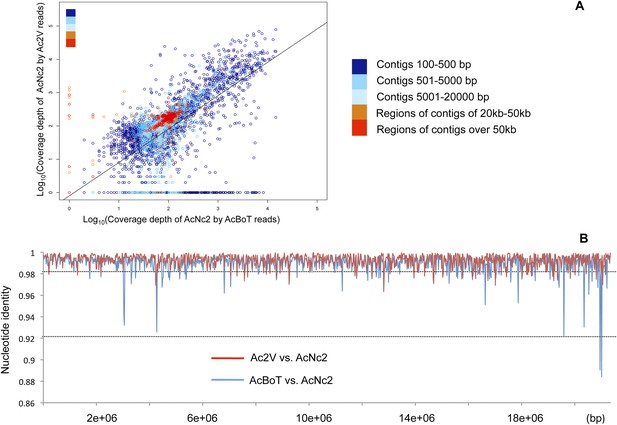
Comparison of A. candida races using alignments of Illumina reads against the AcNc2 assembly.
(A) Positive correlation between the depths of coverage of the reference assembly (AcNc2) by the Ac2V and AcBoT reads. For the reference contigs less than 20 kb, the mean coverage was calculated across the whole contig length and log-transformed. For the contigs over 20 kb, the mean coverage was calculated for the sliding window of 20 kb and log-transformed. Y-axis shows the log-transformed depth of the reference coverage by the Ac2V reads; X-axis shows the log-transformed depth of the reference coverage by the AcBoT reads. (B) Nucleotide identity amongst the homologous genomic regions of Ac2V, AcBoT and AcNc2. The mean identity was calculated for the sliding window of 20 kb.
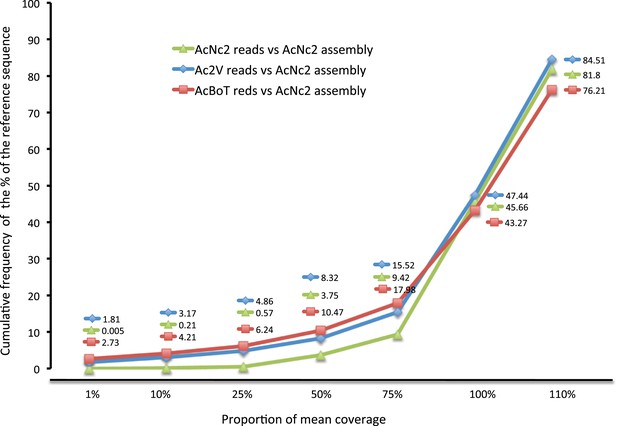
Coverage of the reference assembly (AcNc2) by Ac2V and AcBoT.
Proportion of mean coverage is a measure relative to the depth of coverage of the reference reads mapped backed to the reference assembly. The proportion of AcNc2 assembly not covered by AcBoT and/or Ac2V reads (≤10% of the mean coverage) was 3.17% (971,030 bp) in Ac2V and 4.21% (1,285,877 bp) in AcBoT.
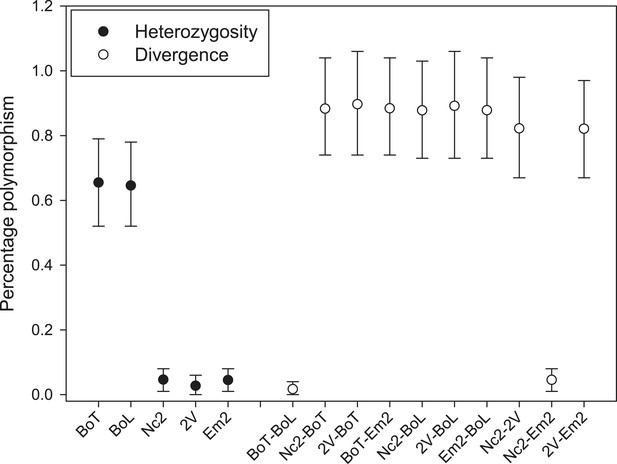
Nucleotide polymorphism within and between A. candida isolates.
Mean (±5–95%CI) polymorphism expressed as the percentage observed heterozygote sites (solid symbols) and percentage nucleotide divergence (open symbols) at contig 1. Confidence intervals were calculated using a bootstrap of contig 1 after removal of indels. Isolates infecting the same host plant (i.e., AcBoT-AcBoL and AcEm2- AcNc2) show little nucleotide divergence, which indicates that they are genotypically almost identical (i.e., diverged by less than 0.05%). Nevertheless, the Brassica oleracea infecting race (AcBoT and AcBoL) possess a relatively high heterozygosity compared to the isolates of the Arabidopsis thaliana infecting race. Moreover, most of this heterozygous polymorphism is shared (low nucleotide divergence) and presence of the majority of heterozygous sites is consistent with clonal reproduction.
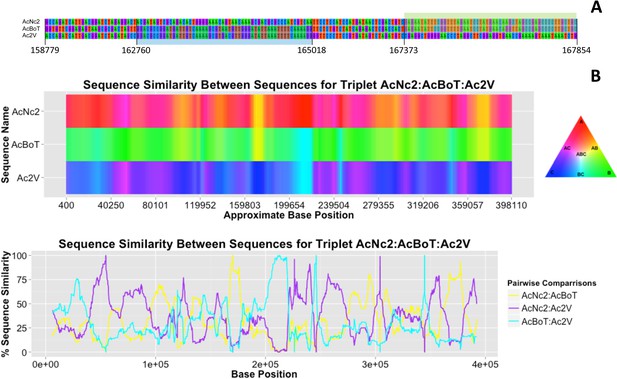
Variation in sequence similarity between races.
(A) Alignment of nucleotides in between positions 158,779 and 167,382 within ‘contig 1’ of three A. candida races (AcNc2, AcBoT and Ac2V) illustrating two recombination blocks coloured blue and green. Both blocks show high sequence similarity between races. Also shown is the sequence divergence in between blocks. Alignment gaps and monomorphic sites have been removed. (B) The sequence similarity at ‘contig 1’ amongst three A. candida races was visualised using the colours of a RBG colour triangular in the software HybRIDS (http://www.norwichresearchpark.com/HybRIDS). Areas where two contigs have the same colour (yellow, purple or turquoise) are indicative of two races sharing the same polymorphisms. The linear plot of the proportion of SNPs shared between the three pairwise comparisons between the races. Shown on the X-axis is the actual base position. The graphs were made in the R package HybRIDS (http://www.norwichresearchpark.com/HybRIDS).
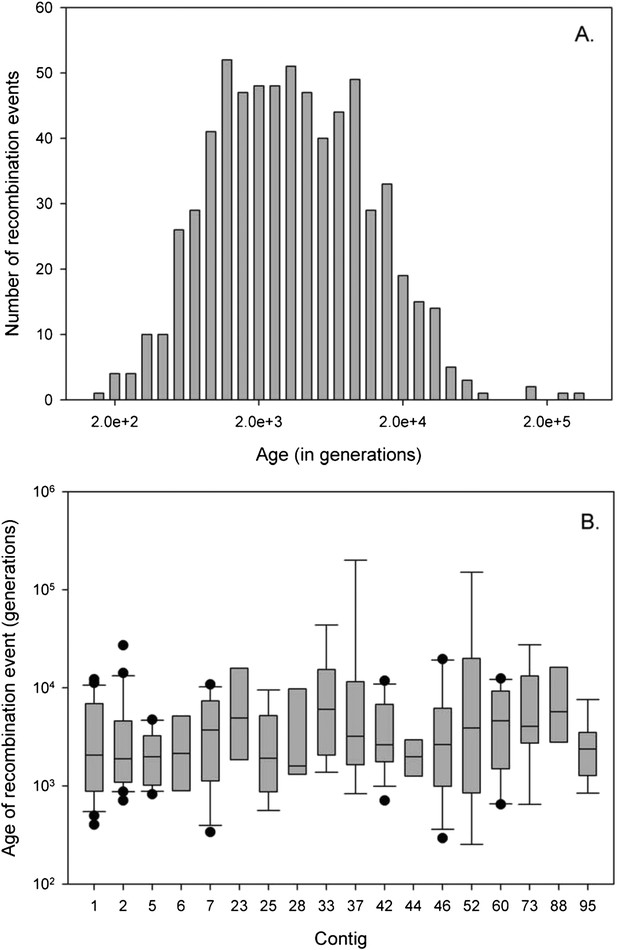
Age of recombination blocks.
(A) Age of the 675 recombination blocks (mutation rate of μ = 10−6) estimated using binomial mass function; (B) Boxplot of the median (plus first nation blocks and third quartile) log-age of recombination events in contigs. Only contigs with eight or more events are shown. There is no significant difference in age of events between contigs (GLM: F22, 233 = 1.06, p = 0.387).
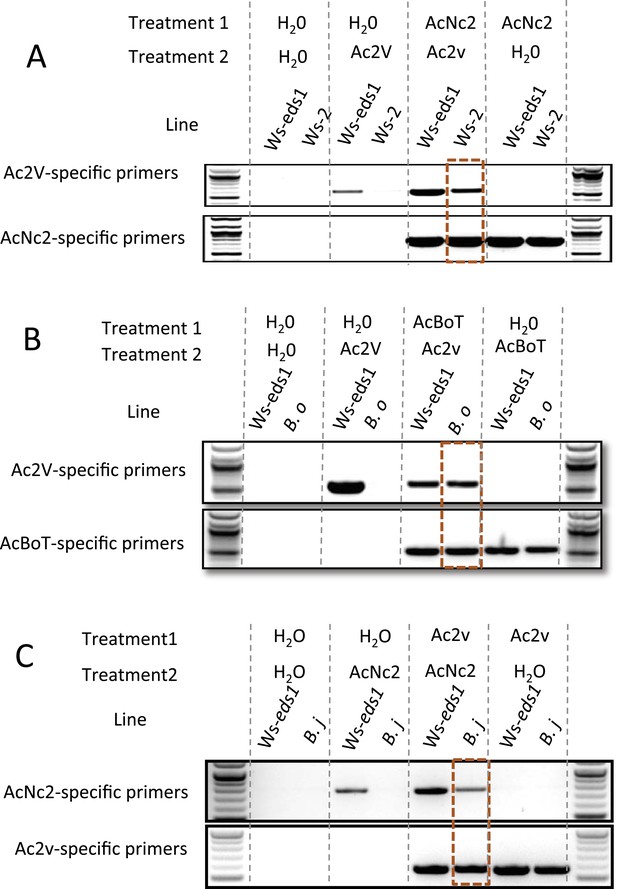
PCR with race-specific markers on DNA prepared from plant tissue generated from co-infection assays.
(A) Co-infection assay of AcNc2 followed by Ac2V onto Ws-eds1 and Ws-2. (B) Co-infection of AcBoT followed by Ac2V onto Ws-eds1 and B. oleracea (B.o). (C) Co-infection of Ac2V followed by AcNc2 onto Ws-eds1 and B. juncea (B.j). Bands highlighted in orange indicate amplification of secondary inoculum on usually non-host plants upon primary inoculation with virulent A. candida. These experiments were repeated multiple times with similar results (see Supplementary file 6).
Tables
Virulence of the A. candida races on different plant host accessions
| A. candida race | Arabidopsis thaliana | Brassica rapa | Brassica junceae | Brassica oleraceae | ||||
|---|---|---|---|---|---|---|---|---|
| + | − | + | − | + | − | + | − | |
| AcNc2 | 137 | 219 | 0 | 1 | 0 | 1 | 0 | 18 |
| AcBoT | 1* | 34 | 0 | 1 | 0 | 2 | 15 | 0 |
| Ac2V | 1*,† | 107 | 1‡ | 4‡ | 6‡ | 0‡ | 0 | 2 |
-
+ Host-pathogen compatible interactions (number of susceptible accessions).
-
− Host-pathogen incompatible interactions (number of resistant accessions).
-
*
A. thaliana Ws-eds1 (enhanced disease susceptibility) mutants were susceptible to all tested A. candida races.
-
†
In the laboratory conditions, the cotyledons of the A. thaliana accession Ws-3 were found to be susceptible to the Ac2V (Cooper et al., 2008).
-
‡
Data from (Rimmer et al., 2000) incorporated; in this study, one cultivar B. rapa (CrGC1-18, rapid-cycling accession) was infected by Ac2V race and four other tested cultivars (‘Torch’, ‘Colt’, ‘Horizon’, ‘Reward’) were incompatible with Ac2V race. All analysed cultivars of B. juncea (CrGC4-1S, ‘Burgonde’, ‘Domo’, ‘Cutlass’) were susceptible to Ac2V.
Summary of the A. candida AcNc2, AcEm2, AcBoT, AcBoL and Ac2V genome assemblies
| AcNc2 | AcEm2 | AcBoT | AcBoL | Ac2V | |
|---|---|---|---|---|---|
| Number of contigs | 5212 | 11,581 | 11,929 | 11,143 | 12,210 |
| N50 length (bp) | 41,078 | 29,326 | 14,673 | 14,953 | 24,005 |
| N50 number | 231 | 284 | 584 | 581 | 353 |
| Mean contig length (bp) | 6610 | 2668 | 2781 | 2998 | 2770 |
| Assembly size (bp) | 34,454,169 | 33,409,146 | 33,184,526 | 33,409,856 | 33,823,601 |
| GC content (%) | 43.19% | 43.09% | 43.15% | 43.15% | 43.11% |
| CEGMA gene space coverage (%)* | 93.55% | 92.74% | 93.55% | 93,13% | 92.34% |
| Average genome coverage | 160 | 150 | 140 | 140 | 200 |
| Repeat content (%) | 17.4% | NA | NA | NA | NA |
| Predicted genes | 10,907 | NA | NA | NA | NA |
-
*
Completeness of the gene space in the different genome assemblies was estimated using CEGMA pipeline; the presence of over 90% of core eukaryotic genes in the assembly serves as an indication of a overall complete gene space.
Additional files
-
Supplementary file 1
List of Arabidopsis thaliana and Brassica spp. accessions assayed in virulence tests with Albugo candida race type isolates AcBoT, Ac2V and AcNc2.
- https://doi.org/10.7554/eLife.04550.012
-
Supplementary file 2
List of Arabidopsis thaliana accessions assayed in virulence tests with Albugo candida isolates AcNc2 and Ac2v.
- https://doi.org/10.7554/eLife.04550.013
-
Supplementary file 3
Polymorphisms between Albugo candida race genomic regions verified with Sanger sequencing.
- https://doi.org/10.7554/eLife.04550.014
-
Supplementary file 4
List of the predicted recombination blocks generated by the RDP3 software.
- https://doi.org/10.7554/eLife.04550.015
-
Supplementary file 5
Details of race-specific primers.
- https://doi.org/10.7554/eLife.04550.016
-
Supplementary file 6
Details of co-infections assays.
- https://doi.org/10.7554/eLife.04550.017





