General principles for the formation and proliferation of a wall-free (L-form) state in bacteria
Figures
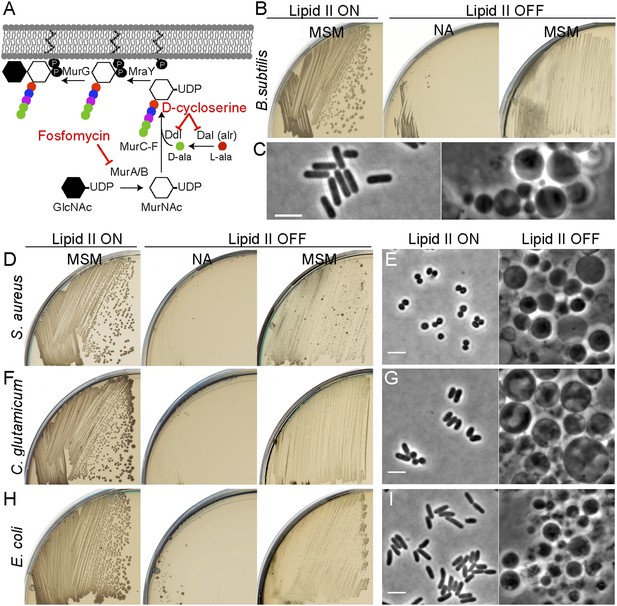
Inhibition of PG precursor synthesis induces L-form proliferation in bacteria.
(A) Schematic model of peptidoglycan (PG) precursor (lipid II) synthesis in bacteria and its inhibition by the antibiotics fosfomycin (FOS) and D-cycloserine (DCS). MurA, inhibited by the antibiotic FOS, and MurB catalyze the transformation of uridine diphosphate-N-acetylglucosamine (UDP-GlcNAc) into UDP-N-acetylmuramic acid (UDP-MurNAc). The racemase Dal and the D-alanine ligase Ddl, both of which are inhibited by the antibiotic DCS, are required to generate D-Ala-D-Ala. This is incorporated into the UDP-MurNAc-pentapeptide, requiring MurC, MurD, MurE, and MurF enzymes. UDP-MurNAc-pentapeptide is transferred to undecaprenyl pyrophosphate by MraY, and the addition of GlcNAc is catalyzed by MurG to form lipid II. (B) Growth of Bacillus subtilis strain LR2 (ispA Pxyl-murE-B) streaked on L-form supporting medium (MSM) or nutrient agar (NA) plates in the presence (lipid II ON) or absence (lipid II OFF) of 0.5% xylose. (C) Phase contrast microscopy of B. subtilis LR2 cells grown on MSM plates in the presence (left) or absence (right) of 0.5% xylose. (D–I) Growth on plates (D, F, H) and corresponding phase contrast microscopy (E, G, I) of bacterial strains Staphylococcus aureus ATCC2913 (D, E), Corynebacterium glutamicum ATCC13032 (F, G), and Escherichia coli MG1655 (H, I). (D, F, H) The different bacterial strains were streaked on MSM or NA plates in the absence (lipid II ON) or presence (lipid II OFF) of the antibiotics FOS (D, H) or DCS (F). (E, G, I) Phase contrast microscopy of the different bacterial cells grown on MSM plates in the absence (left) or presence (right) of the antibiotics FOS (E, I) or DCS (G). Scale bars, 3 μm.
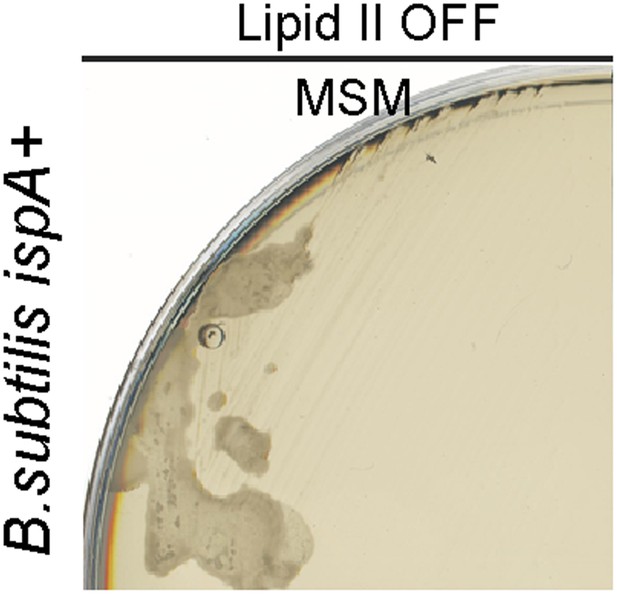
Bacillus subtilis L-form growth requires an additional mutation in a gene such as ispA.
Growth of B. subtilis strain BS115 (Pxyl-murE-B) streaked on L-form supporting medium (MSM) in the absence (lipid II OFF) of 0.5% xylose.
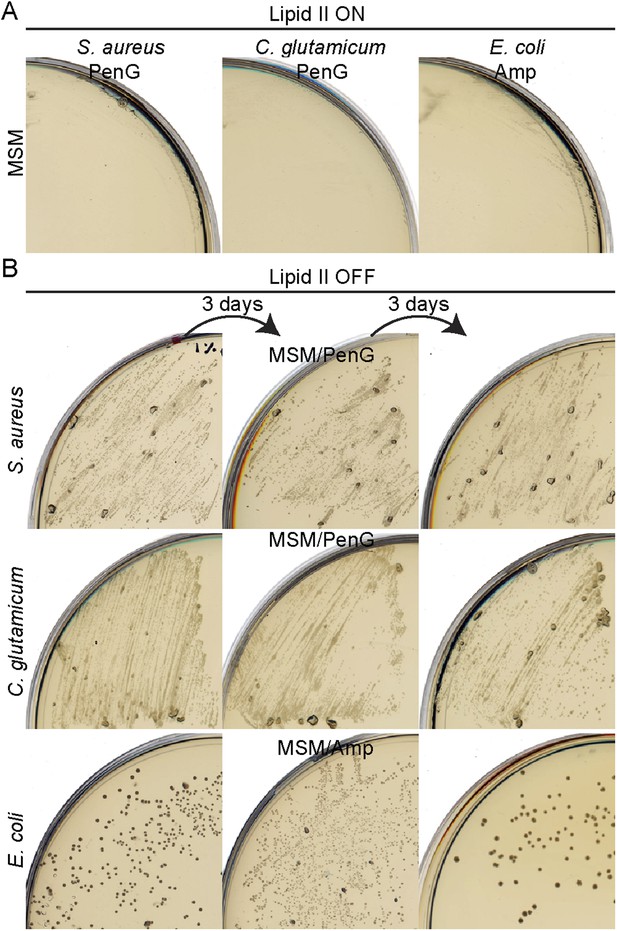
Bacterial L-forms proliferate on β-lactams.
(A) Growth of Staphylococcus aureus (left), Corynebacterium glutamicum (middle), and Escherichia coli (right) walled strains streaked on L-form supporting medium (MSM) in the presence of penicillin G (S. aureus and C. glutamicum) or ampicillin (E. coli). (B) Growth of S. aureus (top), C. glutamicum (middle), and E. coli (bottom) L-forms streaked on MSM with the antibiotics fosfomycin (S. aureus and E. coli) or D-cycloserine (C. glutamicum) in the presence of penicillin G (S. aureus and C. glutamicum) or ampicillin (E. coli). The different L-form strains were streaked several times under the same conditions every 3 days (left to right).
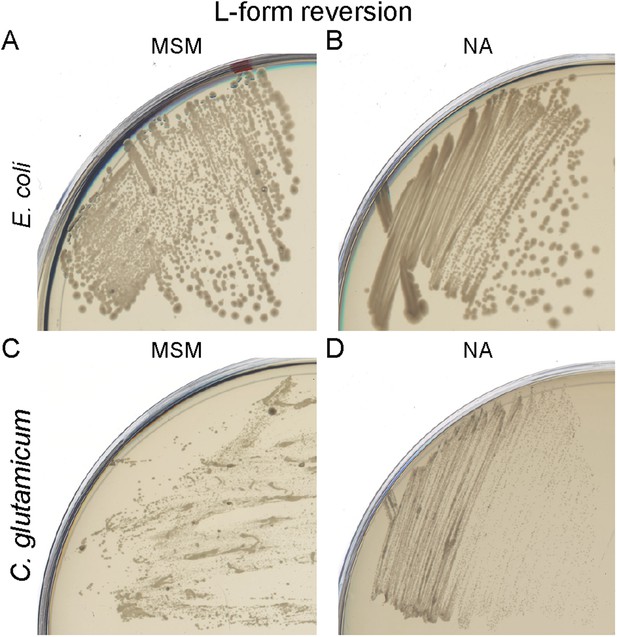
Bacterial L-form cell wall reversion.
(A) Cell wall reversion on L-form supporting medium (MSM) plates of L-forms of Escherichia coli strain TB28 grown on MSM plates containing fosfomycin and ampicillin. (B) Growth of the E. coli L-form reverted strain TB28 from panel (A) on nutrient agar (NA). (C) Cell wall reversion on MSM plates of L-forms of Corynebacterium glutamicum grown on MSM plates with D-cycloserine and penicillin G. (D) Growth of the C. glutamicum L-form reverted strain from panel (C) on NA.
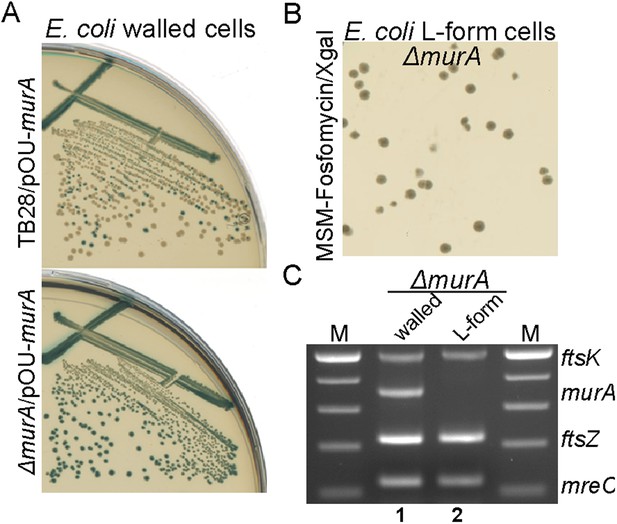
E. coli L-forms proliferate independently of the peptidoglycan cell wall machinery.
(A) Growth of the Escherichia coli strains TB28 (top) and RM345 (ΔmurA, bottom) containing the unstable plasmid pOU82-Amp-murA streaked on nutrient agar plates in the presence of X-gal. (B) L-form colonies of the E. coli strains RM345 (ΔmurA, pOU82-Amp-murA) on L-form supporting medium (MSM) plates in the presence of fosfomycin (FOS) and X-gal, after several repeated streakings on MSM plates in the presence of FOS. (C) Multiplex PCR of the ftsK, murA, ftsZ, and mreC genes from genomic DNA of the E. coli strains RM345 grown in the walled (1) or L-form (2) states, obtained from the strains in panel (B). M represents the 100 bp DNA ladder.
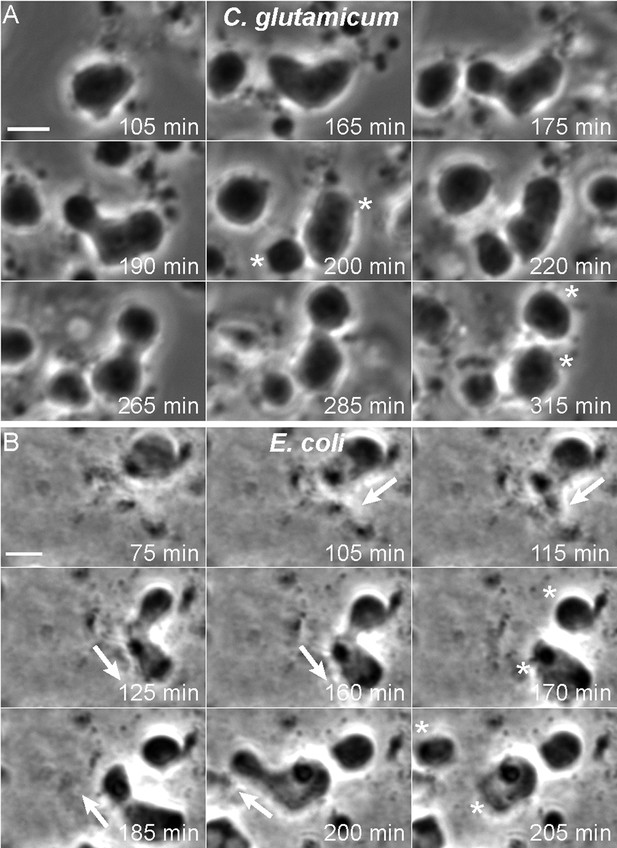
Mode of cell division of E. coli and C. glutamicum L-forms.
(A, B) Corynebacterium glutamicum L-form strain grown in nutrient broth (NB)/L-form supporting medium (MSM) with D-cycloserine (A), and Escherichia coli L-form strain RM345 (ΔmurA) grown on nutrient agar/MSM (B), were observed by time lapse phase contrast microscopy. Elapsed time (min) is shown in each panel. Scale bars, 3 μm. Arrows represent the direction of protrusion formation and the asterisks (*) the daughter cells after division. See also Videos 1–4.
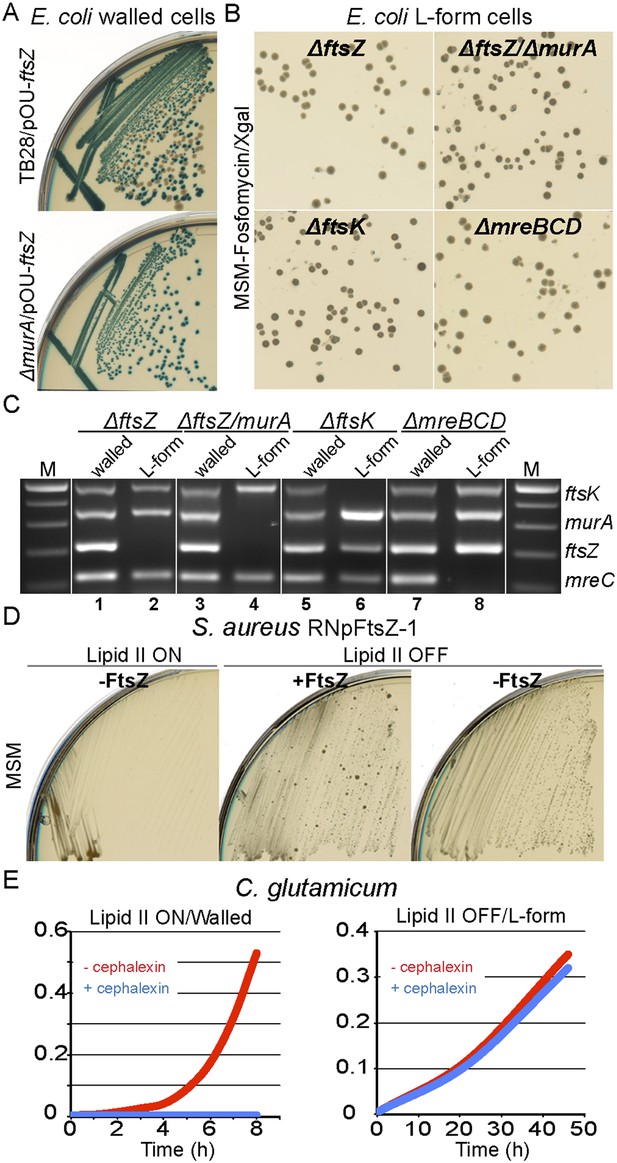
Bacterial L-forms proliferate independently of the cell division machinery.
(A) Growth of the Escherichia coli strains TB28 (top) and RM349 (ΔftsZ, bottom) containing the unstable plasmid pOU82-Amp-ftsZ streaked on nutrient agar plates in the presence of X-gal. (B) L-form colonies of the E. coli strains RM349 (ΔftsZ, pOU82-Amp-ftsZ, top left), RM350 (ΔmurA, ΔftsZ, pOU82-Amp-ftsZ, pSK122-Cm-murA, top right), RM61 (ΔftsK, pSK122-Cm-ftsK, bottom left), and RM359 (ΔmreBCD, pHM82-Kn-mreBCD) on L-form-supporting medium (MSM) plates in the presence of fosfomycin (FOS) and X-gal, after several repeated streakings on MSM plates in the presence of FOS. (C) Multiplex PCR of the ftsK, murA, ftsZ, and mreC genes from genomic DNA of the E. coli strains RM349 (1, 2), RM350 (3, 4), RM61 (5, 6), and RM359 (7, 8) grown in the walled (1, 3, 5 and 7) or L-form (2, 4, 6 and 8) states obtained from the strains in panel (B). M represents the 100 bp DNA ladder. (D) Growth of the Staphylococcus aureus strain RNpFtsZ-1 (erm-pSPAC-ftsZ, Pinho and Errington, 2003) streaked on MSM plates in the absence (lipid II ON, left) or presence (lipid II OFF, middle and right) of FOS, with (+FtsZ, middle) or without (−FtsZ, left and right) isopropyl β-d-1-thiogalactopyranoside. (E) Growth profiles of Corynebacterium glutamicum in MSM with (L-form state; right, lipid II OFF) or without (walled state; left, lipid II ON) D-cycloserine, and in the absence (red) or presence (blue) of cephalexin.
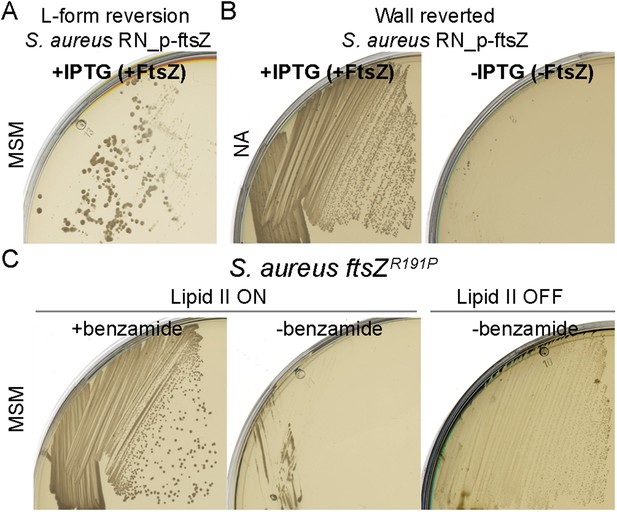
Staphylococcus aureus L-forms proliferate in the absence of the cell division machinery.
(A) Cell wall reversion on L-form supporting medium (MSM) plates with isopropyl β-d-1-thiogalactopyranoside (IPTG) (+FtsZ) of L-forms of S. aureus strain RNpFtsZ-1 grown on MSM plates with fosfomycin (FOS) and without IPTG, obtained from Figure 4D, right. (B) Growth of the S. aureus RNpFtsZ-1 L-form reverted strain from panel (D) on nutrient agar plates with (+FtsZ) or without (−FtsZ) IPTG. (C) Growth of the S. aureus strain ATCC2913 ftsZR191P (Haydon et al., 2008) streaked on MSM plates with (lipid II ON, left and middle) or without (lipid II OFF, right) FOS, and in the presence (left) or absence (middle and right) of benzamide.
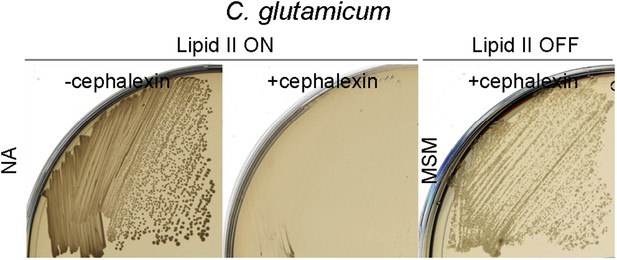
Corynebacterium glutamicum L-forms proliferate in the absence of the cell division machinery.
Growth of the C. glutamicum strain streaked on nutrient agar (NA) (lipid II ON) in the absence (left, −cephalexin) or presence (middle, +cephalexin) of cephalexin, and on L-form supporting medium (MSM) plates with D-cycloserine and cephalexin (right, lipid II OFF, + cephalexin).
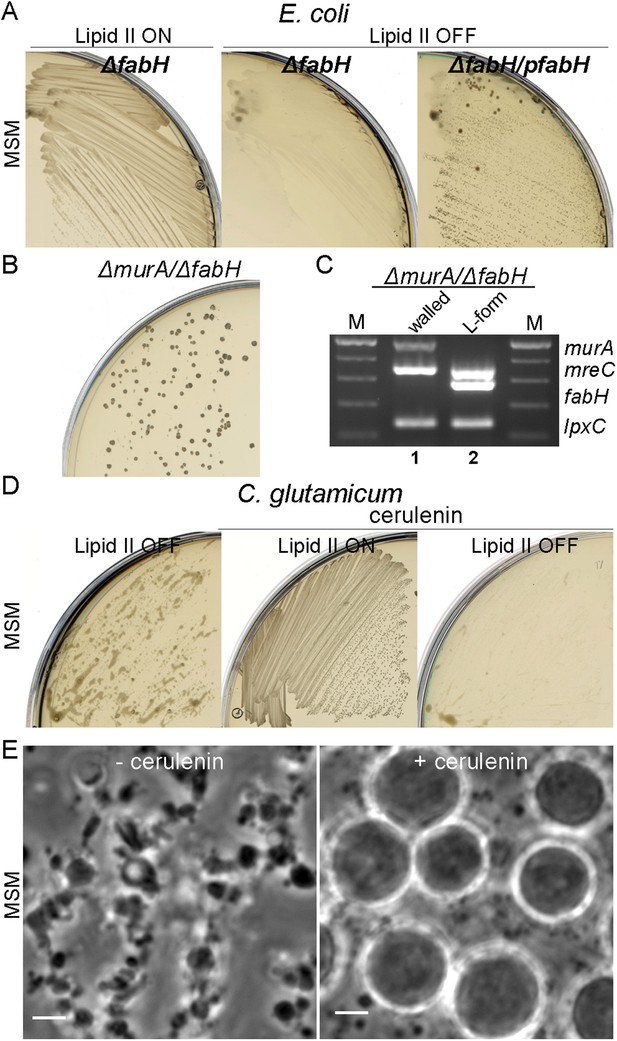
Essential role of fatty acid synthesis in L-forms growth of E. coli and C. glutamicum.
(A) Growth of Escherichia coli strains RM365 (ΔfabH) and RM366 (ΔfabH, pCA24N-fabH) streaked on L-form supporting medium (MSM) in the absence (lipid II ON) or presence (lipid II OFF) of fosfomycin (FOS). (B) L-form colonies of the E. coli strain RM369 (ΔmurA, pSK122-Cm-ftsK, ΔfabH, pOU82-Amp-fabH) on MSM plates after several repeated streakings on MSM plates in the presence of FOS. (C) Multiplex PCR of the genes, murA, fabH, and mreC on genomic DNA of the E. coli strain RM369 grown in the walled (1) or L-form (2) state. Samples obtained from strains in panel (B). M represents the 100 bp DNA ladder. (D) Growth of Corynebacterium glutamicum streaked on MSM in the absence (lipid II ON) or presence (lipid II OFF) of D-cycloserine (DCS), and with (cerulenin) or without (no) 2 μg/ml of cerulenin. (E) Typical images of C. glutamicum L-forms after 16 hr of growth in MSM with DCS in the absence (−cerulenin) or presence (+cerulenin) of 2 μg/ml of cerulenin. Scale bars, 3 μm. See also Videos 5 and 6.
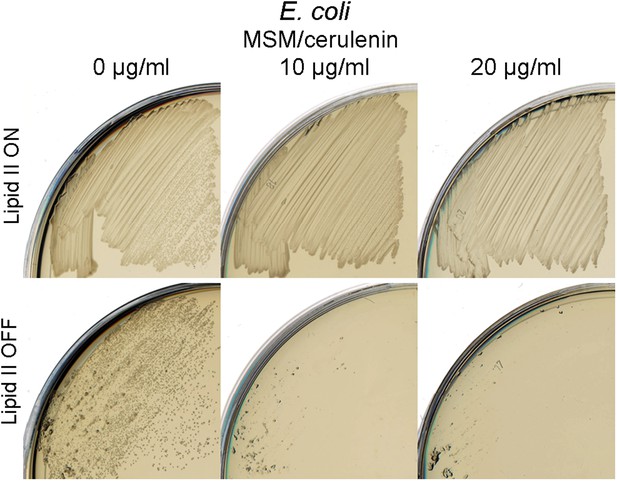
Specific inhibition of Escherichia coli L-forms growth by cerulenin.
Growth of E. coli walled (top) and L-form (bottom) strains streaked on L-form supporting medium (MSM) in the absence (lipid II ON) or presence (lipid II OFF) of fosfomycin with different concentration of cerulenin (0, 10, and 20 μg/ml).
Videos
Time lapse series showing L-form cell growth of Corynebacterium glutamicum growing in nutrient broth (NB)/L-form supporting medium (MSM) with D-cycloserine (DCS), from which the panels in Figure 3A were obtained. Phase contrast images were acquired automatically every 5 min for about 5 hr. Scale bar, 3 μm.
Time lapse series showing L-form cell growth of Corynebacterium glutamicum growing in nutrient broth (NB)/L-form supporting medium (MSM) with D-cycloserine (DCS). Phase contrast images were acquired automatically every 5 min for about 3 hr 30 min. Scale bar, 3 μm.
Time lapse series showing L-form cell growth of Escherichia coli strain RM345 (ΔmurA) growing in nutrient agar (NA)/L-form supporting medium (MSM) from which the panels in Figure 3B were obtained. Phase contrast images were acquired automatically every 5 min for about 4 hr. Scale bar, 3 μm.
Time lapse series showing L-form cell growth of Escherichia coli strain RM345 (ΔmurA) growing in nutrient agar (NA)/L-form supporting medium (MSM). Phase contrast images were acquired automatically every 5 min for about 4 hr. Scale bar, 3 μm.
Time lapse series showing L-form cell growth of Corynebacterium glutamicum growing in nutrient broth (NB)/L-form supporting medium (MSM) with D-cycloserine (DCS) in the absence of 2 μg/ml of cerulenin. Phase contrast images were acquired automatically every 5 min for about 16 hr. Scale bar, 3 μm.
Time lapse series showing L-form cell growth of Corynebacterium glutamicum growing in nutrient broth (NB)/L-form supporting medium (MSM) with D-cycloserine (DCS) in the presence of 2 μg/ml of cerulenin. Phase contrast images were acquired automatically every 5 min for about 16 hr. Scale bar, 3 μm.
Tables
General properties of bacterial L-forms
| Bacterial L-form strain | Bacillus subtilis | Staphylococcus aureus | Corynebacterium glutamicum | Escherichia coli |
|---|---|---|---|---|
| Mode of induction | MurE-B repression | FOS | DCS | FOS |
| Secondary mutation required | Yes | n.d. | n.d. | n.d. |
| Timing of induction | 24 hr | 3 days | 48h | 3 days |
| Growth condition | Solid/liquid | Solid | Solid/liquid | Solid |
| Cell wall reversion | Yes | Yes | Yes | Yes |
| Cell division machinery | Not essential | Not essential | Not essential | Not essential |
| Mode of cell proliferation | Vesicles blebbing, fission, tubulation | n.d. | Vesicles blebbing, fission, tubulation | Vesicles blebbing, fission, tubulation |
| References | Leaver et al., 2009, Mercier et al., 2013, Kawai et al., 2014 |
-
DCS: D-cycloserine; FOS: fosfomycin; n.d.: not determined.
Bacterial strains and plasmids used in this study
| Strain | Relevant genotype | Reference |
|---|---|---|
| Bacillus subtilis | ||
| Bs115 | 168CA ΩspoVD::cat Pxyl-murE ΩamyE::(tet xylR) | (Leaver et al., 2009) |
| LR2 | Bs115 xseB* (frameshift 22T > −)a | (Mercier et al., 2013) |
| Escherichia coli | ||
| TB28 | MG1655 ΔlacIZYA | (Bernhardt and de Boer, 2003) |
| ND101 | fstK::Kn pSC101-fstK | F-X Barre Lab unpublished |
| RM61 | TB28 ΔftsK::kan pSK122-ftsK | This study |
| RM345 | TB28 ΔmurA::kan pOU82-murA | This study |
| RM349 | TB28 ΔftsZ::kan pOU82-ftsZ | This study |
| RM350 | TB28 ΔftsZ pOU82-ftsZ ΔmurA::kan pSK122-murA | This study |
| RM359 | TB28 ΔmreBCD::cat pTK549 | (Kruse et al., 2005) |
| RM365 | TB28 ΔfabH::kan | (Baba et al., 2006) |
| RM366 | RM365 pCA24N-fabH | This study |
| RM369 | TB28 ΔfabH pOU82-fabH ΔmurA::kan pSK122-murA | This study |
| Staphylococcus aureus | ||
| WT | ATCC29213 | Laboratory collection |
| S. aureus ftsZR191P | ATCC29213 ftsZR191P | (Haydon et al., 2008) |
| RNpFtsZ-1 | RN4220 Pspac-ftsZ erm | (Pinho and Errington, 2003) |
| Corynebacterium glutamicum | ||
| WT | ATCC13032 | Laboratory collection |
| Plasmid | Relevant genotype | Reference/origin |
|---|---|---|
| Escherichia coli | ||
| pCA24N-fabH | lacIq pT5-lac-fabH cat | (Kitagawa et al., 2005) |
| pOU82 | R1-replicon, bla lacZYA | (Gerdes et al., 1985) |
| pOU82-murA | R1-replicon, bla lacZYA murA | This study |
| pOU82-ftsZ | R1-replicon, bla lacZYA ftsZ | This study |
| pOU82-fabH | R1-replicon, bla lacZYA fabH | This study |
| pTK549 | R1-replicon, kan Pmre-mreBCD | (Kruse et al., 2005) |
| pSK112 | F-replicon, cat lacZYA | F-X Barre Lab unpublished |
| pSK112-ftsK | F-replicon, cat lacZYA ftsK | F-X Barre Lab unpublished |
| pSK112-murA | F-replicon, cat lacZYA murA | This study |
-
bla: b-lactamase; cat: chloramphenicol; erm: erythromycin; lacZ: β-galactosidase; kan: kanamycin; tet: tetracyclin.





