The transforming growth factor beta signaling pathway is critical for the formation of CD4 T follicular helper cells and isotype-switched antibody responses in the lung mucosa
Figures
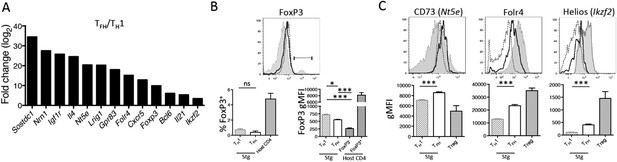
TGF-β-associated gene expression signature in Tfh cells.
(A) Bar graph shows a selected set of genes upregulated in d8 LCMV-specific Stg PSGL1lo Ly6Clo Tfh cells relative to PSGL1hi Ly6Chi Th1 cells isolated and sorted from the spleen as measured using Illumina DNA microarrays (Marshall et al., 2011) that have been described to be induced by TGF-β or associated with Treg cells (Hill et al., 2007). (B) Representative histogram plot (top) shows amount of intracellular FoxP3 in total host splenic CD4 T cells (shaded gray) and LCMV-specific Th1 (hatched line) and Tfh (black line) Stg CD4 T cells from the spleen at day 8 p.i. Region gated identifies FoxP3+ nTregs. Bar graphs (bottom) depict the cumulative frequency (left) of FoxP3+ CD4 T cells or gMFI averages (right) of the indicated CD4 T cell populations. (C) Expression of the indicated Treg-associated proteins in (A) was compared between LCMV-specific Th1 (hatched line) and Tfh cells (black line), and FoxP3+ Treg cells gated on total host CD4 T cells (shaded gray) from the spleen at day 8 p.i. Histogram plots (top) are representative examples of individual mice and bar graphs (bottom) depict the gMFI averages of each protein in the indicated CD4 T cell populations. Graphs in B and C are representative of one of five independent experiments (n = 4–5 mice/group/experiment). *p < 0.05, ***p < 0.0005.
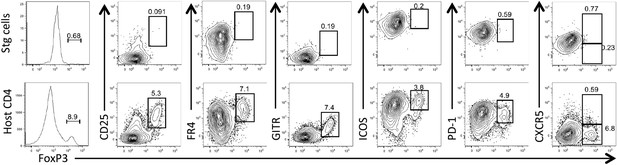
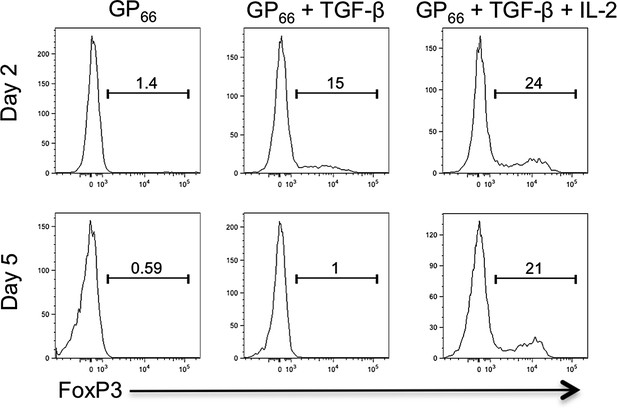
LCMV-specific Stg CD4 T cells do not form canonical regulatory T cells nor T follicular regulatory cells.
1 × 104 Stg CD4 T cells were adoptively transferred into congenic C57BL/6 recipient mice, which were infected with LCMV Armstrong the following day. Eight days p.i., splenocytes were assessed for intracellular FoxP3 expression and the other indicated proteins. Top row depicts LCMV-specific Stg CD4 T cells, while the bottom row is gated on the host-derived CD4 T cells. Data depict an individual mouse representative of more than three independent experiments with 10+ total mice.
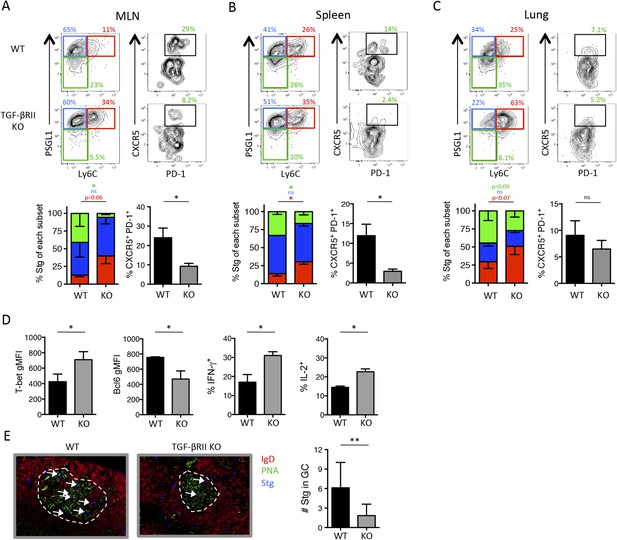
Direct TGF-β is required for influenza-specific Tfh differentiation.
2 × 105 CD44lo TGF-βRII+/+ CD4-cre+ (WT) or TGF-βRIIf/f CD4-cre+ (KO) Stg cells were adoptively transferred into C57BL/6 congenic recipients infected with WSN-GP33/66 the following day. (A–D) On day 8 p.i, Stg cells in the MLN, spleen, and lung were stained with antibodies against the indicated proteins to distinguish Th1 and Tfh cells. Cells were also stimulated with GP66-peptide for 6 hr to assess IFN-γ and IL-2 production by intracellular cytokine staining and flow cytometry. (E) Stg cells (blue, highlighted by white arrows) located within MLN PNA+ GCs (green) were assessed using immunofluorescent microscopy and their numbers were enumerated using Imaris software. Graphs in A–D are representative of one of five independent experiments (n = 4–5 mice/group/experiment). Panel E shows representative microscopy images and the cumulative data from two independent experiments with 8 total mice/group were graphed. *p < 0.05, **p < 0.005, and colored asterisks correspond to the color in the stacked graphs.
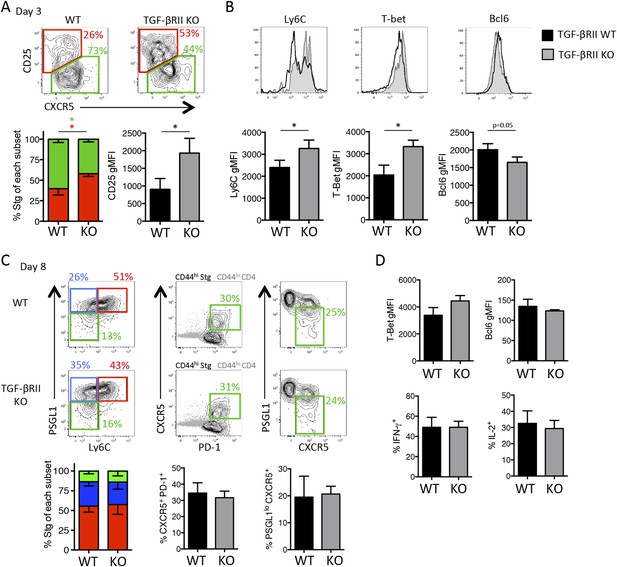
Direct TGF-β restricts anti-viral TH1 precursor formation but does not impact overall effector CD4 T cell differentiation during LCMV.
(A) Stg chimeric mice (1 × 106 CD44lo TGF-βRII+/+ CD4-cre+ (WT) or TGF-βRIIf/f CD4-cre+ (KO)) were infected with LCMV Armstrong and 3 days p.i., spleens were collected and analyzed by flow cytometry for the indicated proteins. (B–D) Stg chimeric mice (1 × 104 CD44lo TGF-βRII+/+ CD4-cre+ (WT) or TGF-βRIIf/f CD4-cre+ (KO)) were infected with LCMV Armstrong and the phenotype of Stg cells was assessed 8 days p.i. Data are representatives of five independent experiments with 15–20 total mice/group. *p < 0.05.
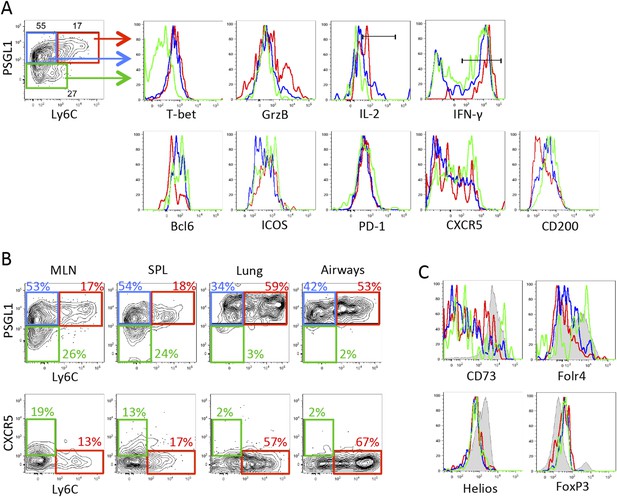
Expression of PSGL1 and Ly6C distinguish between influenza-specific Th1 and Tfh CD4 T cells.
2 × 105 Stg CD4 T cells were adoptively transferred into congenic C57BL/6 recipient mice, which were infected with influenza WSN-GP33/66 i.n. the following day. 8 days p.i., lymphocytes in the MLN were assessed for the indicated proteins. (A and C) Representative example of Stg cells in the MLN. In the histograms, red lines denote PSGL1hi Ly6Chi Stg cells, blue is PSGL1hi Ly6Clo Stg cells, and green is gated on PSGL1lo Ly6Clo Stg cells. (B) Stg cells from the MLN, spleen, lung, and airways. (C) includes FoxP3+ Tregs in the shaded gray histograms. Data depict an individual mouse representative of more than 30 total mice from 10+ independent experiments.
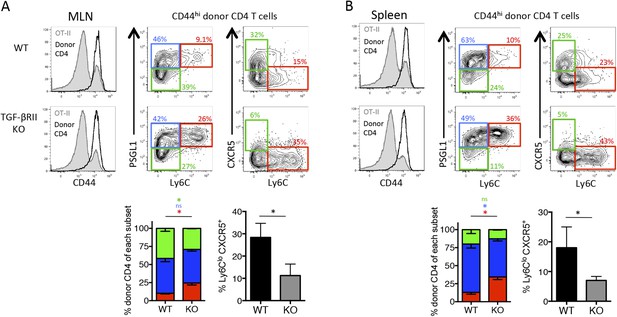
Direct TGF-β signaling is required for polyclonal influenza-specific Tfh differentiation.
5 × 106 CD44lo Thy1.2+ CD4 T cells from TGF-βRIIf/f Lck-cre− (WT) or TGF-βRIIf/f Lck-cre+ (KO) mice were adoptively transferred into congenic Thy1.1+ OT-II TCR transgenic mice and infected with WSN-GP33/66 the following day. 14 days p.i., host OT-II (Thy1.1+) and donor (Thy1.2+) CD4 T cells in the MLN (A) and spleen (B) were assessed for expression of CD44 (to distinguish activated T cells, see histogram plots left) and CXCR5, PSGL1, and Ly6C to distinguish Tfh and Th1 attributes. FACS plots are from representative mice and bar graphs are representative of one of four independent experiments (n = 4–5 mice/group/ experiment). *p < 0.05 and colored asterisks correspond to the color in the stacked graphs.
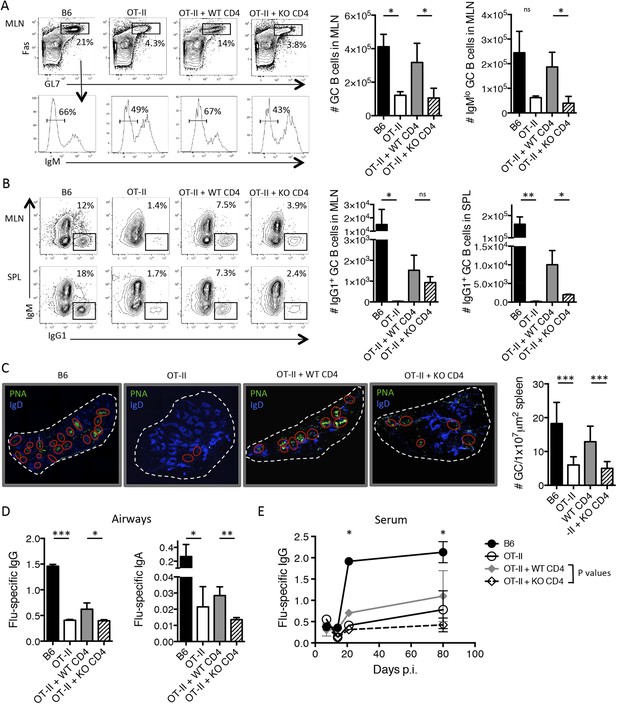
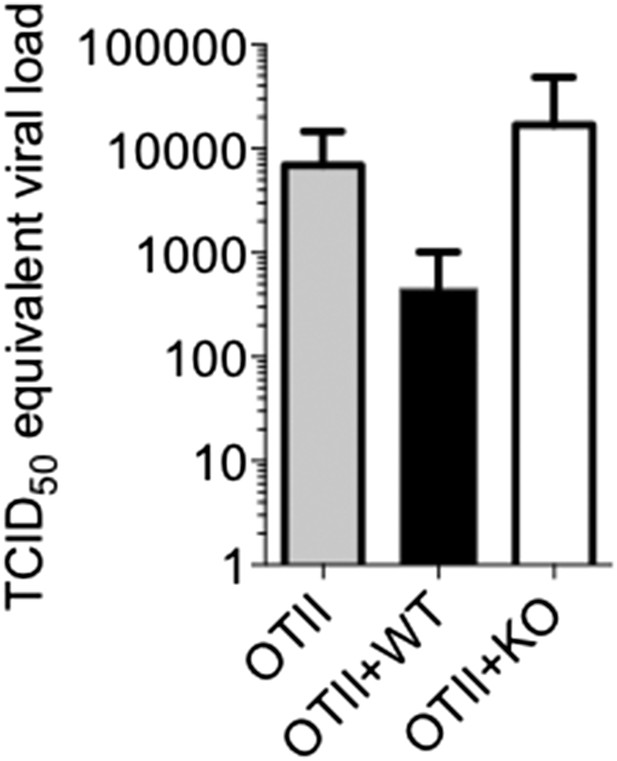
T cell-directed TGF-β is required for GC B cell and isotype-switched antibody responses during influenza infection.
5 × 106 CD44lo Thy1.2+ CD4 T cells from TGF-βRIIf/f Lck-cre− (WT) or TGF-βRIIf/f Lck-cre+ (KO) mice were adoptively transferred into congenic Thy1.1+ OT-II TCR transgenic mice and infected with WSN-GP33/66 the following day. 14 days p.i., GC B cells (Fas+, GL7+ IgM−) (A) and IgG1+ GC B cells (B) in the MLN and spleen (SPL) were assessed by flow cytometry (left plots) and enumerated in bar graphs (right). (C) Splenic PNA+ GCs (green, highlighted by red ellipses) were assessed by immunofluorescent microscopy of frozen sections and the numbers of GC/tissue section was calculated using Imaris software. (D–E) Influenza-specific IgG and IgA were measured from bronchoaviolar lavage fluid (BAL) by ELISA at day 10 p.i. (D) or longitudinally at the indicated time points (E). Data in panels A–B are representative of four independent experiments (n = 3–5 mice/group/experiment). The bar graphs in panels C–D show cumulative data from two independent experiments (n = 3–5 mice/group/experiment), the images in panel C are from representative mice of these cohorts. *p < 0.05, **p < 0.005, ***p < 0.0005.
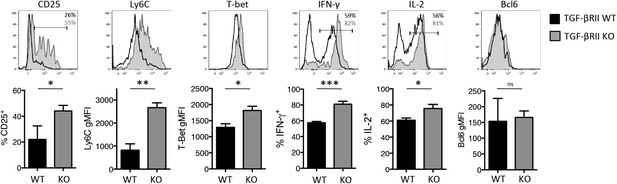
Direct TGF-β suppresses early influenza-specific Th1 precursor formation in the lung-draining MLN.
2 × 106 TGF-βRII+/+ CD4-cre+ (WT) or TGF-βRIIf/f CD4-cre+ (KO) Stg cells were adoptively transferred into C57BL/6 congenic recipients that were infected with WSN-GP33/66 the following day. On day 4–5 p.i., CD44hi Stg cells in the MLN were assessed for the indicated proteins (CD25, Ly6C, T-bet, IFN-γ, IL-2, Bcl6) by flow cytometry. Cells were stimulated with PMA and ionomycin for 4 hr to assess IFN-γ and IL-2. Bar graphs are representative of three independent experiments (n = 3–4 mice/group/experiment). *p < 0.05, **p < 0.005, ***p < 0.0005.
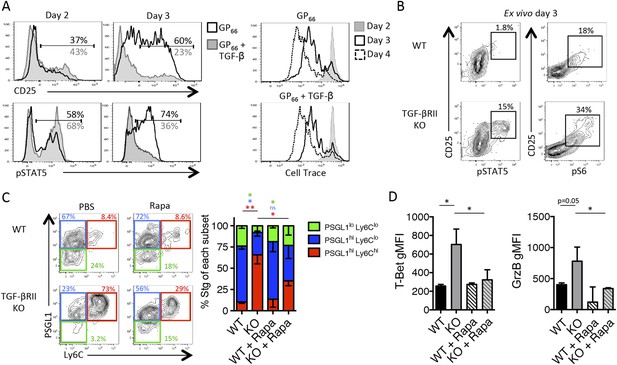
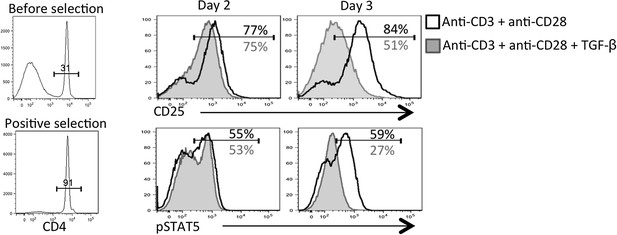
TGF-β restricts IL-2 responsiveness and insulates early Tfh progenitor cells from mTOR signaling.
(A) Stg CD4 T cells were labelled with Cell Trace dye and cultured in vitro with 0.1 μM GP66 peptide ± 10 ng/ml TGF-β and stained for surface expression of CD25 (top) or restimulated with IL-2 to assess pSTAT5 (bottom). Data are representative of four independent experiments. (B) Stg chimeric mice (1 × 106 CD44lo TGF-βRII+/+ CD4-cre+ (WT) or TGF-βRIIf/f CD4-cre+ (KO)) were infected with LCMV Armstrong and 3 days p.i., spleens were fixed immediately in 2% PFA and stained with antibodies to measure surface expression of CD25, Ly6C, and intracellular phosphorylation of STAT5 (pSTAT5) and pS6 directly ex vivo. Data are representative of three independent experiments including 3–5 total mice/group. (C–D) 2 × 105 CD44lo TGF-βRII+/+ CD4-cre+ (WT) or TGF-βRIIf/f CD4-cre+ (KO) Stg cells were adoptively transferred into congenic C57BL/6 recipients infected with WSN-GP33/66 the following day. Mice were treated with PBS or 75 mg/kg rapamycin i.p. daily. On day 8 p.i, Stg cells in the MLN were assessed for expression of PSGL1 and Ly6C (C) or T-bet or GzmB (D). Data are representative of three independent experiments encompassing a total of 9–15 mice/group. *p < 0.05, **p < 0.005 and colored asterisks correspond to the color in the stacked graphs.