The contrasting phylodynamics of human influenza B viruses
Figures
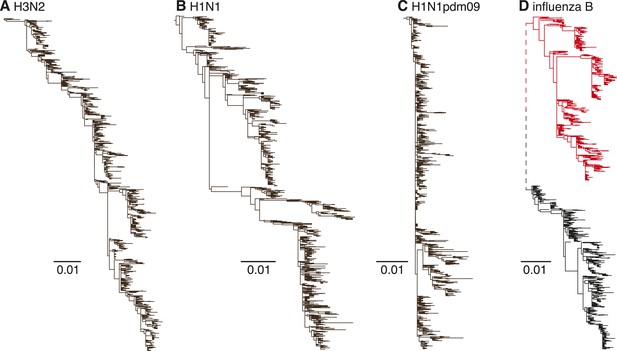
Evolutionary dynamics of human influenza A and influenza B Victoria and Yamagata viruses.
Evolution of the HA genes of influenza A H3N2 virus, 2002–2013, (A), H1N1 virus, 1998–2009 (B), H1N1pdm09 virus, 2009–2013 (C), and influenza B Yamagata (red) and Victoria (black) lineage viruses, 2002–2013 (D). All phylogenetic trees were generated using approximately 1200 randomly selected full-length gene sequences sampled during 12 years.
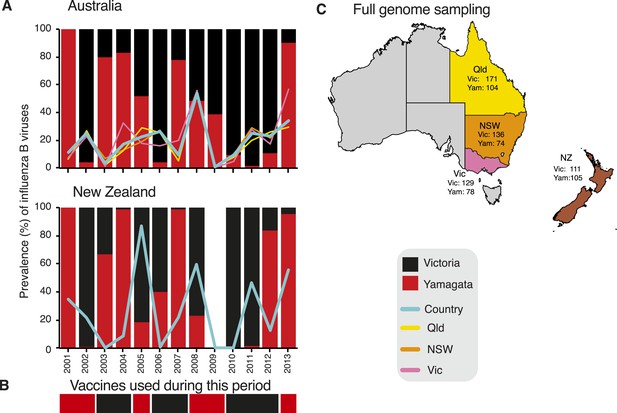
Influenza B virus lineages in Australia and New Zealand, 2001–2013 and source of full genomes.
Percentage prevalence of influenza B viruses isolated from the three eastern Australian states and New Zealand (A). Coloured lines represent the proportion of influenza viruses typed as influenza B in each country (blue) and each of the eastern Australian states; Queensland (yellow), New South Wales (orange), and Victoria (pink). Bars represent the percentage prevalence of Victoria (black) and Yamagata (red). Data based on National Notifiable Diseases Surveillance system (NNDSS) for Australia and Environmental Science and Research (ESR) for New Zealand. The lineage of representative influenza B virus strains used in the trivalent influenza vaccine during these years in both countries (B). Excluding the years 2003 and 2009, influenza B viruses represented on average 24.6% (range 9.5–53.7%) and 31.5% (range 0.5–86.9%) of laboratory confirmed influenza viruses from Australia and New Zealand, respectively. The percentage of circulating influenza viruses that were influenza B was significantly lower in 2003 (AUS, 3.4%) and 2009 (AUS, 0.8%) than in other years, due to the dominance of a new H3N2 variant (A/Fujian/412/2002-like) in 2003 and the emergence of the H1N1 pandemic in 2009. Source of full genomes of Victoria and Yamagata viruses (C).
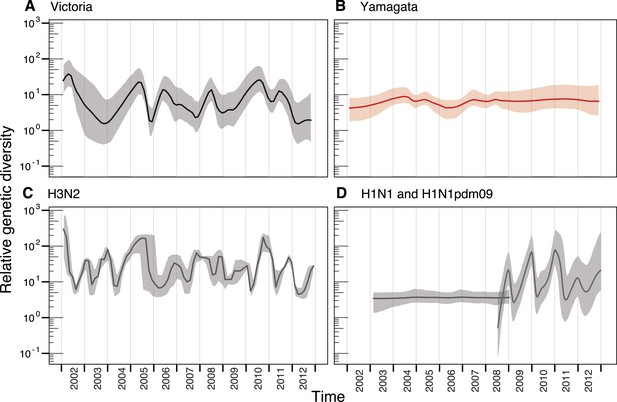
Population dynamics of genetic diversity in Australia and New Zealand.
The relative genetic diversity of the HA segments of influenza B Victoria (A), Yamagata (B) and influenza A H3N2 (C), and H1N1 2003–2008 and H1N1pdm09 2009–2013 viruses (D), isolated in Australia and New Zealand using the Gaussian Markov Random Field (GMRF) model.
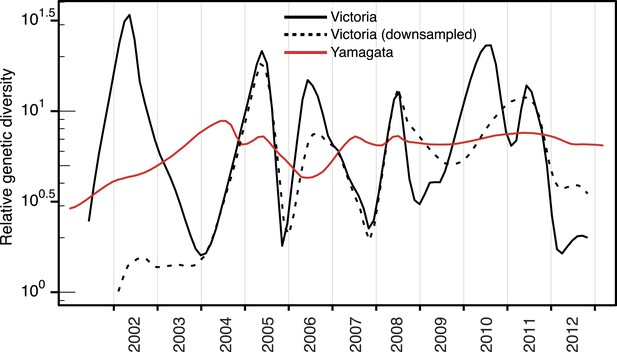
Effect of sampling on the population dynamics of Influenza B virus.
Relative genetic diversity of the Victoria (black) and Yamagata (red) lineages estimated using the Gaussian Markov Random Fields (GMRF) Skyride model (as in Figure 3), using a subsampled Victoria data set, in which, the number of Victoria lineage viruses was randomly reduced to match the size of Yamagata for that year.
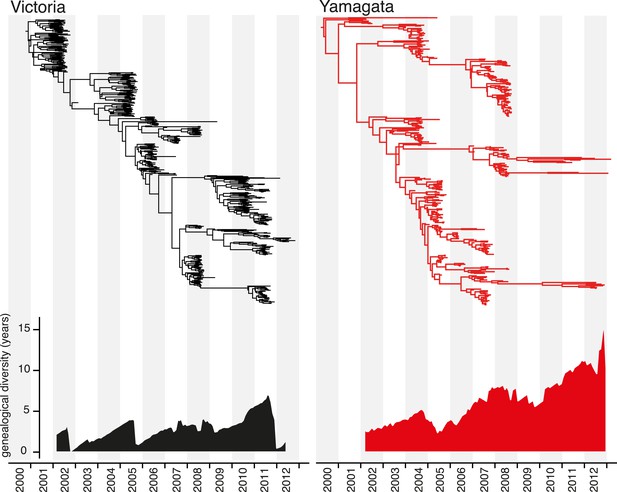
Evolution of the hemagglutinin genes of influenza B viruses.
Phylogenetic relationship of the HA genes of influenza B Victoria (black) and Yamagata (red) lineage viruses inferred using the uncorrelated lognormal relaxed clock model. Genetic diversity through time was estimated by averaging the pairwise distance in time between random contemporaneous samples with a 1-month window on the same dated Maximum clade credibility (MCC) trees.
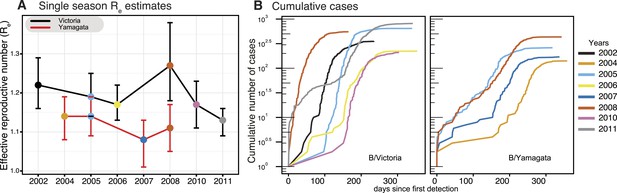
Phylodynamics and cumulative cases of influenza B viruses.
Effective reproductive number (Re) of influenza B Victoria (black) and Yamagata (red) viruses (of the HA data set) estimated for single epidemics (median and 95% highest posterior density (HPD) values) during years with sufficient number of sequences estimated using the BDSIR model (A). The cumulative number of cases from all influenza B virus positive samples for each of these years (B).
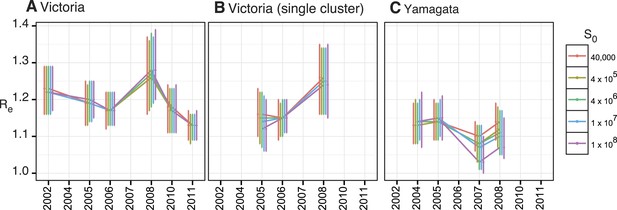
Estimates of Re with various S0 values.
Estimates of effective population size, Re, using various S0 values for all Victoria (A) and Yamagata (C) lineage viruses isolated in Australia and for the largest monophyletic group of Victoria (B) viruses in Australia that clearly represent a single introduction.
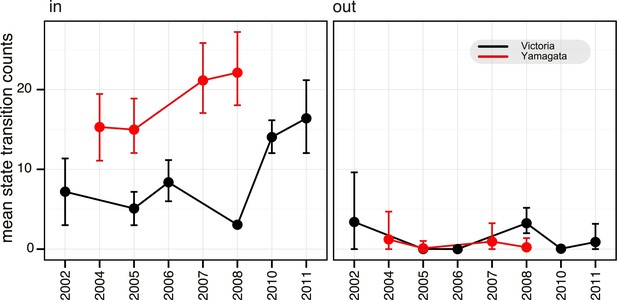
Estimation of migration of influenza B viruses into and out of Australia and New Zealand.
Estimated counts of import and export of Victoria (black) and Yamagata (red) between Australia and New Zealand and rest of the world, using the HA gene data set. Error bars represent the 95% highest posterior density (HPD) values of each point.
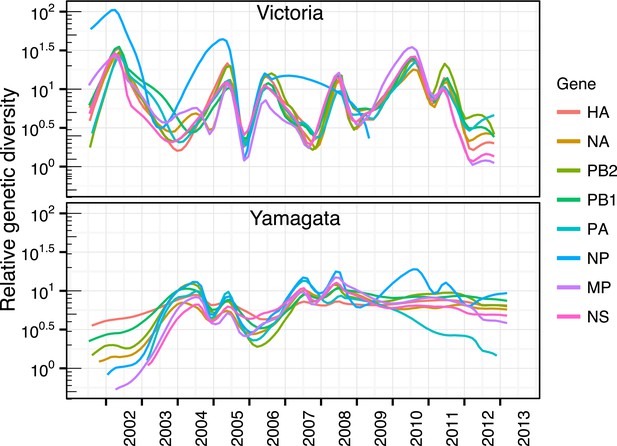
Genome wide evolutionary dynamics—relative genetic diversity.
Relative genetic diversity of each gene segments of Victoria (black) and Yamagata (red) lineages estimated using the Gaussian Markov Random Fields (GMRF) Skyride model (as in Figure 3).
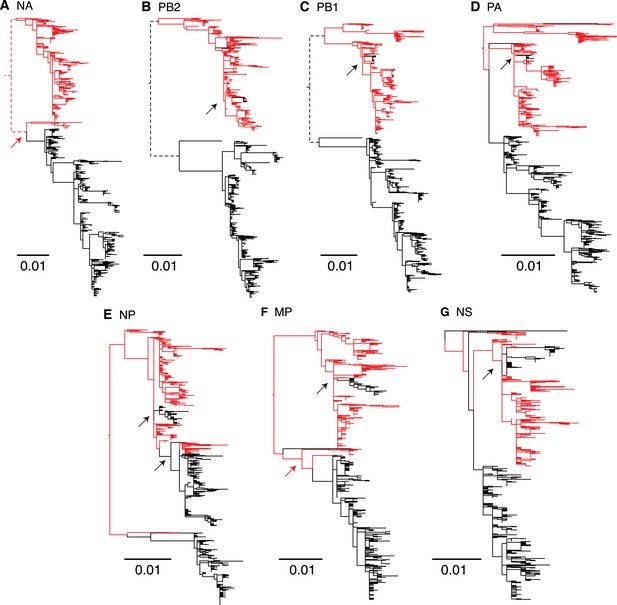
Genome wide evolutionary dynamics—reassortment.
Evolutionary relationships of neuraminidase (A), polymerase basic 2 (B), polymerase basic 1 (C), polymerase acidic (D), nucleoprotein (E), matrix (F), and non-structural (G) genes of Victoria and Yamagata lineage viruses inferred using the maximum likelihood analysis of 908 full genome sequences. Lineages are coloured based on the HA lineage: Victoria (black) and Yamagata (red) and arrows highlight inter-lineage reassortment.
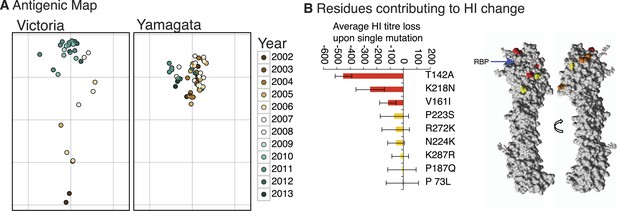
Antigenicity of influenza B viruses.
Antigenic map showing relative antigenic differences of Victoria and Yamagata lineage viruses (circles) measured using the hemagglutinin inhibition (HI) assay for each strain and coloured by year of isolation (A). Residues contributing to HI titer changes (B). Among the nine amino acid changes that we detected between antigenically different Victoria viruses, three changes produced strong HI titer change (>100) (red), 3 medium (≈50) (orange) and 3 low (<20) (yellow). Changes that produced the strongest HI titer change were the closest to the receptor binding pocket (blue arrow), highlighting the significance of their proximity to HI titer change. Amino acids were mapped on previously resolved influenza B virus structure (PDB:4FQM). Detailed HI titer values and reference antigens used are provided in the Dryad source data (Vijaykrishna et al., 2015).
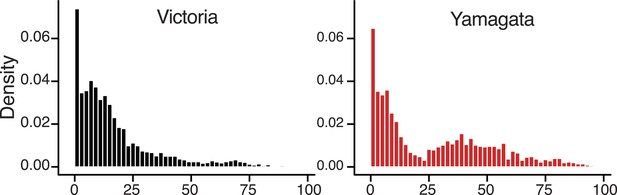
Age distribution of influenza B viruses.
Density of age distribution of influenza B virus positive samples of Victoria (black) and Yamagata (red) lineages, collected from Australia and New Zealand during 2002–2013. Patient age was available for 5260 samples. The age distributions by lineage were compared by histogram using 2-year bins. Also see Table 2 for comparison by age categories and Dryad source data for mean and median age for each year.
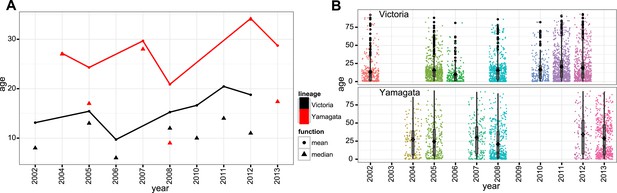
Year-wise age distribution of influenza B viruses.
Mean and median of age distribution of influenza B viruses (A). Box-whisker plot with mean (square) and age distribution of all influenza B viruses cases (jitter plot) are shown for years with greater than 100 samples for either lineage (B).
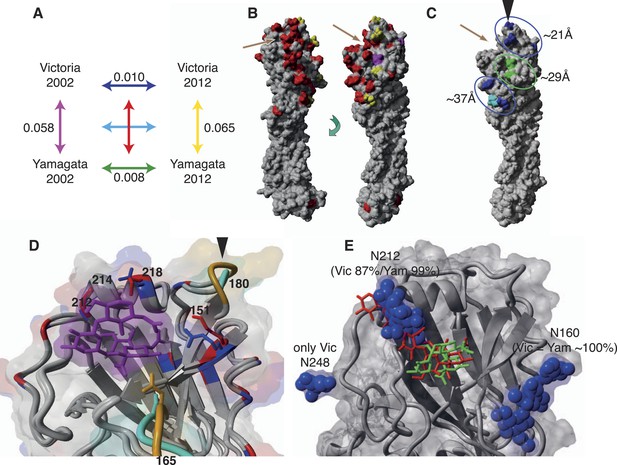
Structural view of the HA showing mutational accumulation and lineage differences.
Amino acid changes observed within and between influenza B virus lineages (A). Arrow colours in (A) correspond to inter- (B) or intra- (C) lineage amino acid changes, based on previously resolved crystal structure (PDB:4FQM). Amino acids in red represent differences between the two lineages that were retained over all sampling years; yellow represents differences that are newly observed in 2012 compared to 2002; and magenta represents changes lost in 2012 compared to 2002. Amino acids in blue and green represent changes that occurred in Victoria and Yamagata viruses between 2002 and 2012, respectively; whereas cyan represents difference between 2002 and 2012 shared between both lineages. These amino acid changes occur in regions that cluster around 21, 29, and 37 Å distant from the RBP (C). Structural differences in RBP among recent Victoria (B/Brisbane/60/2008) and Yamagata (B/Florida/4/2006) strains with a human-like α-2,6 host receptor analogue (magenta) modeled within the viral RBP (D). D was based on crystal structures PDB:4FQM and PDB:4FQJ with side-chains minimized after addition of ligand from PDB:2RFU through superposition. Regions differing in backbone conformation are shown in orange for Victoria and cyan for Yamagata, while conserved regions are shown in gray. Residues with conserved backbone structure but different amino acid side-chains are shown in red for Victoria and blue for Yamagata. Side-chains are shown only for residues within 5 Å of the receptor ligand and differing between the lineages. Structural view of receptor binding pocket with α-2,6- (green) and α-2,3-linked (red) host receptor and glycans (blue) (E). E was based on crystal structure PDB:4FQM, with the addition of ligands from PDB:2RFU and PDB:2RFT through superposition and no minimization. The presence of a glycan on site 212 allows binding only to 2,6-linked receptors, while loss of the glycan allows binding to both α-2,3- and α-2,6-linked receptors. Brown arrows (B and C) indicate relative position of receptor binding pocket (RBP), whereas black arrow heads (C and D) point to site of known antigenic cluster transition (Koel et al., 2013).
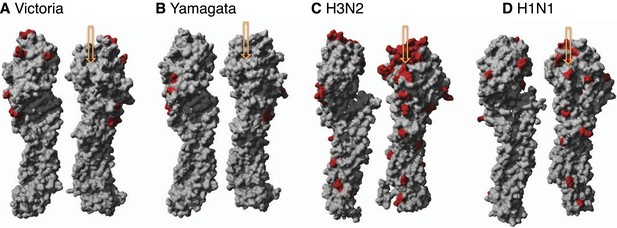
Structural view of mutational drift in influenza A and B viruses.
Amino acid mutations accumulated over 10 years (red) using different rotations of the hemagglutinin monomer structure of influenza B Victoria (2002–2012) (PDB:4FQM) (A), Yamagata (2002–2012) (PDB:4FQM) (B) in comparison to seasonal influenza A H3N2 (1999–2009) (PDB:2YP4) (C), and H1N1 (1997–2007) (PDB:3UBE) (D) viruses. Arrows point to receptor binding pocket.
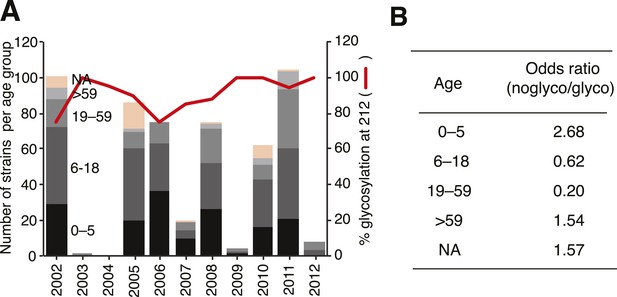
Glycosylation at Asn 212 and correlation with age groups for Victoria viruses.
Yamagata viruses showed five instances of glycosylation loss at 212, compared to 71 instances in Victoria, hence Victoria lineage strains have been analyzed in detail here. Temporal distribution of age groups and glycosylation at 212 for all Victoria strains (A). Summary of odds ratio (OR) for association of glycosylation loss at 212 with the different age groups (B). OR values >1 indicate that it is more likely to find a 212 loss in the respective age group, whereas values <1 indicate that 212 losses are less likely to be found in the respective groups. The following guideline helps judging significance of OR: strong positive association >3; moderate positive association 1.5–3; moderate negative association 0.33–0.66; strong negative association <0.33.
Tables
Nucleotide substitution rates (nucleotide substitutions/site/year) and selection pressures (dN/dS) of influenza B viruses from Australia and New Zealand during 2002–2013
| Mean substitution rates | Branch dN/dS | Site dN/dS | |||||
|---|---|---|---|---|---|---|---|
| Segment* | (95% HPD) | Global dN/dS | Internal | External | Internal/External | No. +ve (sites) | No. −ve |
| Victoria | |||||||
| PB2 | 1.49 (1.28–1.69) | 0.08 (0.07–0.09) | 0.02 | 0.03 | 0.55 | 0 | 373 |
| PB1 | 0.14 (0.12–0.16) | 0.08 (0.07–0.09) | 0.06 | 0.05 | 1.08 | 1 (474) | 402 |
| PA | 1.65 (1.44–1.88) | 0.13 (0.11–0.15) | 0.08 | 0.08 | 1.03 | 1 (700) | 334 |
| HA | 2.00 (1.74–2.57) | 0.19 (0.17–0.22) | 0.12 | 0.09 | 1.37 | 2 (212, 214) | 239 |
| NP | 1.04 (0.76–1.34) | 0.09 (0.07–0.12) | 0.07 | 0.05 | 1.22 | 0 | 49 |
| NA | 2.04 (1.72–2.36) | 0.31 (0.28–0.35) | 0.25 | 0.24 | 1.02 | 6 (46, 73, 106, 145, 146, 395) | 129 |
| MP | 1.44 (1.17–1.70) | 0.06 (0.04–0.09) | 0.00 | 0.02 | 0.01 | 0 | 87 |
| NS | 1.71 (1.38–2.06) | 0.45 (0.38–0.53) | 0.11 | 0.30 | 0.37 | 3 (116, 120, 249) | 13 |
| Yamagata | |||||||
| PB2 | 2.00 (1.74–2.25) | 0.06 (0.05–0.07) | 0.03 | 0.02 | 1.44 | 0 | 443 |
| PB1 | 1.78 (1.56–2.00) | 0.07 (0.05–0.08) | 0.02 | 0.03 | 0.82 | 1 (357) | 392 |
| PA | 1.60 (1.35–1.84) | 0.10 (0.08–0.12) | 0.03 | 0.05 | 0.57 | 0 | 204 |
| HA | 2.01 (1.73–2.29) | 0.13 (0.11–0.16) | 0.07 | 0.07 | 0.98 | 0 | 245 |
| NP | 1.87 (1.65–2.10) | 0.10 (0.08–0.11) | 0.08 | 0.07 | 1.16 | 0 | 308 |
| NA | 2.25 (1.90–2.60) | 0.20 (0.17–0.24) | 0.30 | 0.18 | 1.70 | 1 (295) | 124 |
| MP | 2.20 (1.85–2.55) | 0.05 (0.03–0.07) | 0.05 | 0.02 | 2.08 | 0 | 102 |
| NS | 2.00 (1.66–2.39) | 0.33 (1.66–2.39) | 0.42 | 0.32 | 1.32 | 0 | 30 |
-
*
Analysis was restricted to the non-overlapping regions of M1 and NS1, for the MP and NS segments, respectively.
Age distribution by group
| Victoria | Yamagata | ||||
|---|---|---|---|---|---|
| Age | n | % | n | % | p value* |
| <6 | 1007 | 28.8 | 473 | 26.8 | |
| 6–17 | 1361 | 39 | 402 | 22.7 | |
| ≥18 | 1124 | 32.2 | 893 | 50.5 | |
| Total | 3492 | 100 | 1768 | 100 | <0.0001 |
-
*
Age categories were compared by lineage using a χ2 test.
Summary of evolutionary and epidemiological characteristics of influenza B virus lineages
| Characteristics | Victoria | Yamagata |
|---|---|---|
| Age distribution | younger (mean 16.8, median 11) | older (mean 26.6, median 18) |
| Genetic diversity | strong seasonal changes | weak seasonal changes |
| R (medians) | higher (1.13–1.27) | lower (1.08–1.14) |
| Positive selection | stronger | weaker |
| Antigenic drift | relatively strong | relatively weak |
| Reassortment | high inter-sublineage reassortment, with lower intra-sublineage reassortment | low inter-sublineage reassortment, with greater intra-sublineage reassortment |
| Receptor binding preference | α-2,3- and α-2,6-linked sialic acid | mainly α-2,6 linked sialic acid |





