Angiopoietin-like proteins stimulate HSPC development through interaction with notch receptor signaling
Figures
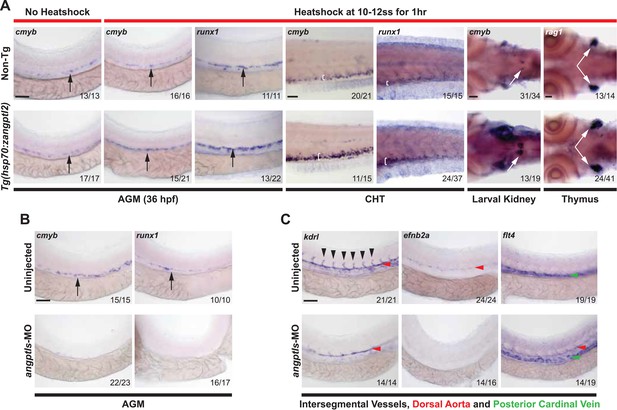
Angptls are sufficient and required for definitive hematopoiesis.
(A) Heatshocked Tg(hsp70:zangptl2) embryos have increased cmyb- and runx1-positive HSPCs in the AGM (black arrows, 36hpf), CHT (white brackets, 3 days post-fertilization, dpf), larval kidney (white arrows, 4dpf) and rag1-positive T-cells in the thymus (white arrows, 5dpf) compared to control no heatshock or non-Tg siblings. (B) Angptls-MO morphants (2 ng) had decreased cmyb- and runx1-positive HSPCs (black arrows) in the AGM at 36hpf and (C) severe disruption to vascular development with loss of kdrl-positive ISVs (black arrowheads), loss of arterial efnb2a and ectopic expression of venous flt4 in the DA (red arrowheads) in addition to PCV (green arrowheads) at 28hpf. Scale bars: 50 μm.
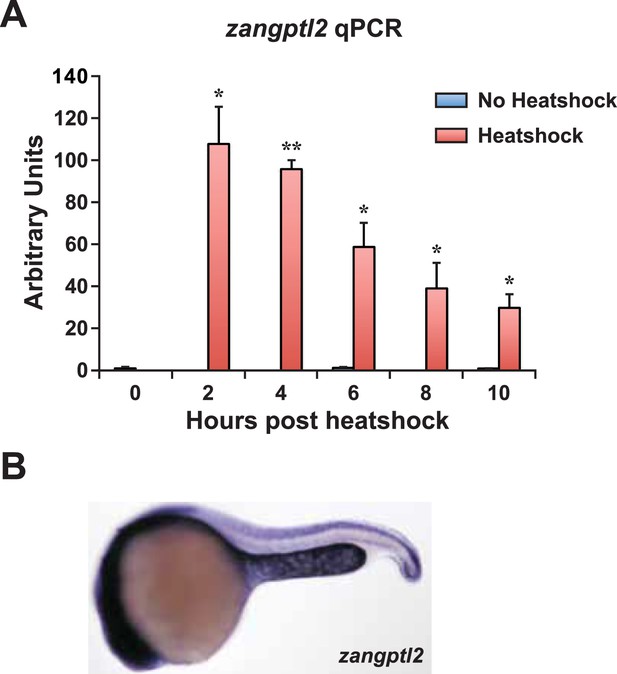
Zangptl2 overexpression in Tg(hsp70:zangptl2) embryos and endogenous zangptl2 expression.
(A) qPCR analysis of zangptl2 mRNA levels in Tg(hsp70:zangptl2) embryos that have been heatshocked for 1 hr and collected at the indicated times post-heatshock. Heatshocked embryos (red bars) overexpressed zangptl2 mRNA at least 100-fold in excess compared to non-heatshocked siblings (blue bars). Error bars denote S.E.M., *p < 0.05, **p < 0.01 compared to 0 hr, one way ANOVA. (B) WISH of endogenous zangptl2 at 23hpf (the highest of all timepoints observed) is mostly restricted in the yolk sac extension, spinal cord, and head region.
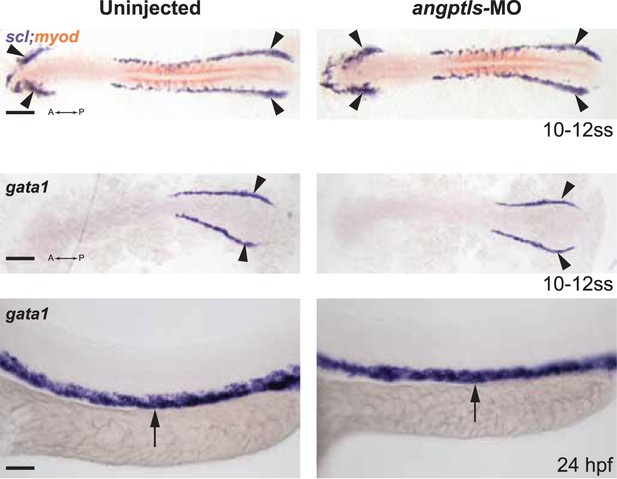
Angptls-MO morphants had no defect in primitive hematopoiesis.
Top panels: somite matched embryos are scored by double staining with myod (orange, staining somite boundaries) and scl (purple, for early blood and vascular progenitor cells in the anterior (A) and posterior (P) bilateral stripes of the lateral plate mesoderm (LPM), black arrowheads, 10–12 ss). Middle and bottom panels: gata1-positive proerythroblast formation was also unaffected in these morphants in the posterior LPM (black arrowheads, 10–12 ss) and intermediate cell mass (black arrows, 24hpf). Scale bars: 200 μm (for top and middle panels); 50 μm (for bottom panels).
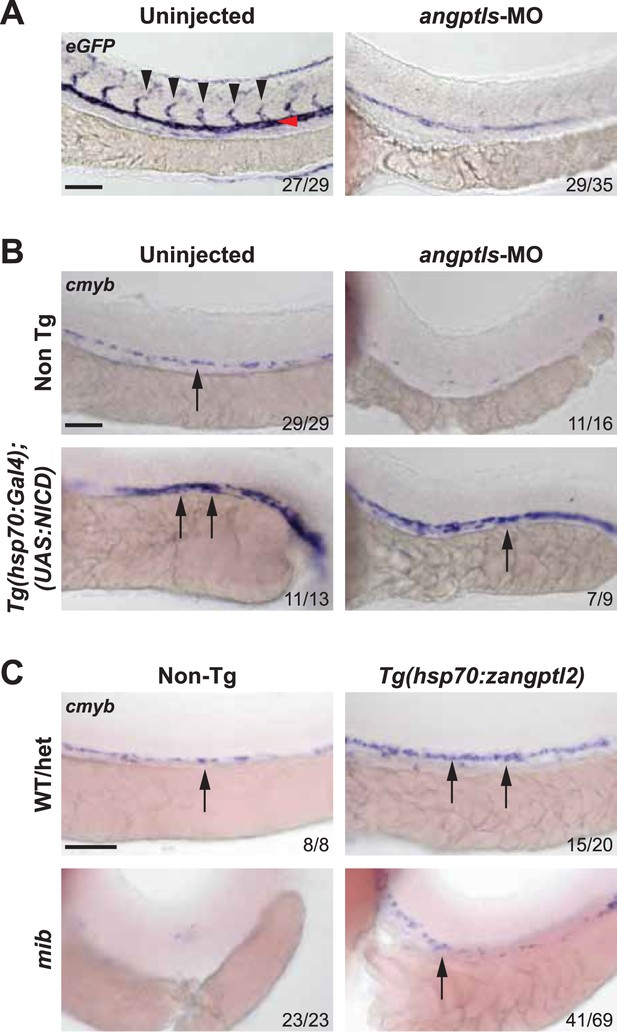
Angptls genetically interact with notch.
(A) Notch eGFP Tg reporter embryos injected with angptls-MO (2 ng) had decreased notch activity in ISV (black arrowheads) and DA (red arrowhead) at 28hpf. (B) Overexpression of constitutively active notch in angptls-MO injected (2 ng) Tg(hsp70:Gal4;UAS:NICD) can restore cmyb-positive HSPCs (arrows) at 36hpf after heatshock. (C) Mib had no cmyb-positive HSPCs (arrows) at 36hpf compared to WT or het siblings. Overexpression of zangptl2 in mib by crossing Tg(hsp70:zangptl2) into mib can restore this defect upon heatshock. Scale bars: 50 μm.
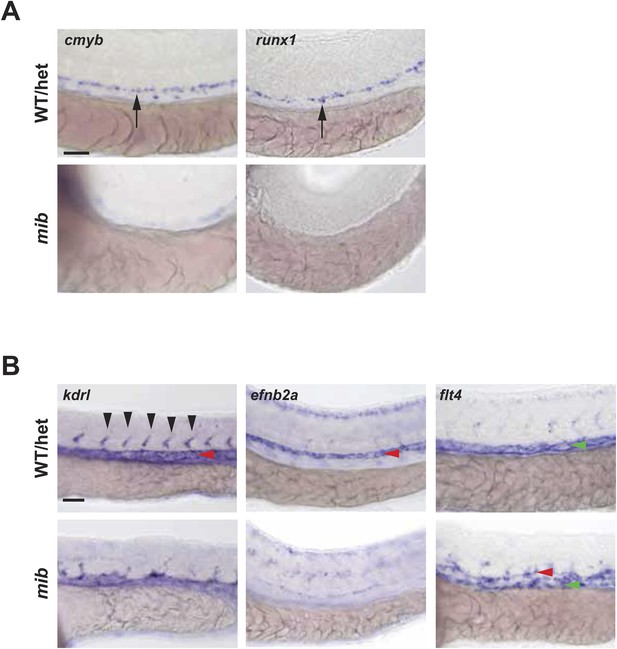
Notch mutant mib has hematopoietic and vascular defects.
(A) Mib has no cmyb- or runx1-positive HSPCs (arrows) in the AGM at 36hpf. (B) Mib embryos also display vascular defects including disorganized ISV sprouting (black arrowheads), loss of arterial marker, efnb2a in the DA (red arrowheads), and ectopic expression of the venous marker, flt4 in the DA in addition to the PCV (green arrowheads) at 28hpf. Scale bars: 50 μm.
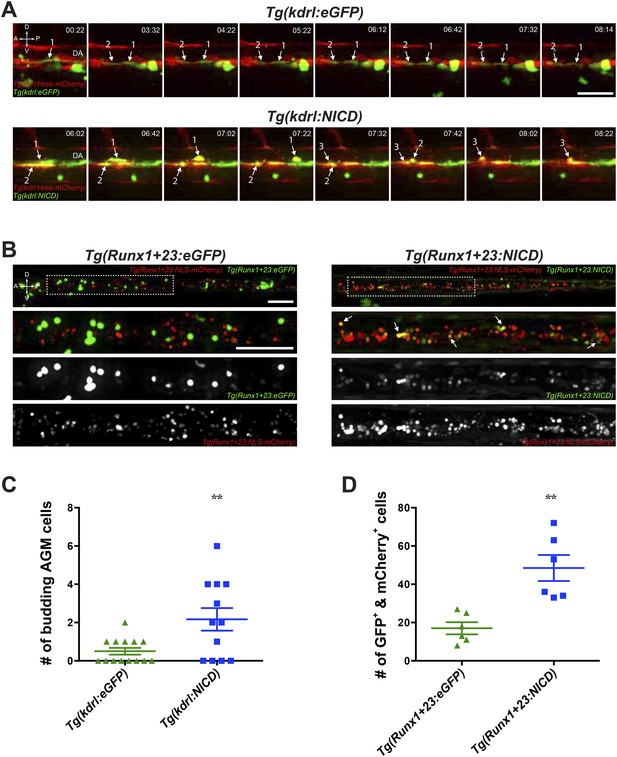
Notch cell autonomously increase definitive hematopoiesis.
(A) Time-lapse sequence (hours:minutes post-28hpf) of HSPCs emerging from the ventral wall of the DA. Hemogenic endothelial cells (red) that have incorporated the injected transgene (green) are marked with numbers and white arrows. Each injected embryo was scored for 24 hr and tabulated in (C). Scale bar: 25 μm. (B) Still images of the CHT from 72hpf embryos. Runx1-positive HSPCs (red) that have incorporated the injected transgene (green) were scored for double positivity (yellow, examples marked by white arrows) and tabulated in (D). Boxed areas in the top panels are magnified and split into double or single fluorescent panels below. Scale bars: 50 μm. A: Anterior, P: Posterior, D: Dorsal; V: Ventral. Error bars denote S.E.M., **p < 0.01, compared to eGFP injected controls, Student's t test.
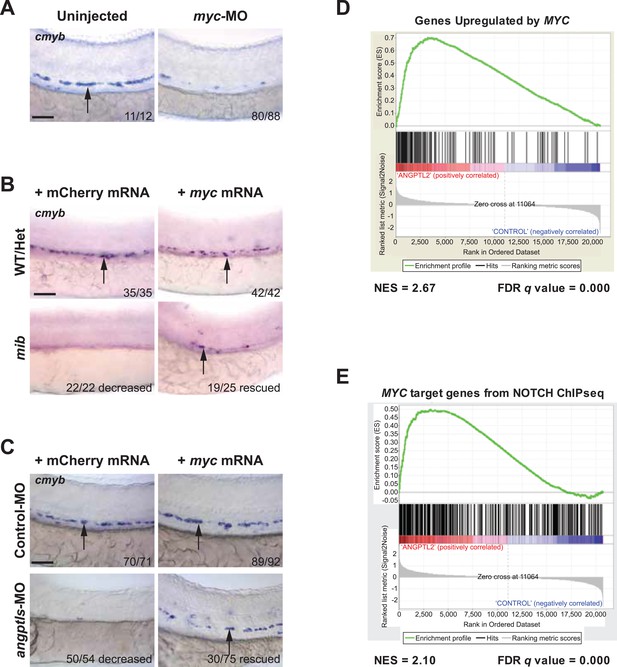
Myc is downstream of angptl and notch signaling.
(A) Myc MO (6 ng) injected embryos had decreased cmyb-positive HSPCs (arrows) at 36hpf. (B) Mib embryos injected with myc mRNA (100 pg) had restored cmyb-positive HSPCs (arrows) at 36hpf compared to siblings injected with control mCherry mRNA (100 pg). (C) Overexpression of myc mRNA (100 pg) in angptls-MO (2 ng) morphants also had restored cmyb-positive HSPCs (arrows) at 36hpf compared to control siblings injected with mCherry mRNA (100 pg). Control-MO: non-targeting MO (2 ng). (D) Representative GSEA plot that positively correlate ANGPTL2-stimulated human CD34+ gene expression and MYC targets. (E) GSEA plot showing significant correlation between MYC target genes from NOTCH ChIPseq and expression data of ANGPTL2-stimulated CD34+ cells. NES, Normalized Enrichment Score, FDR q value, False Discovery Rate q value. Scale bars: 50 μm.
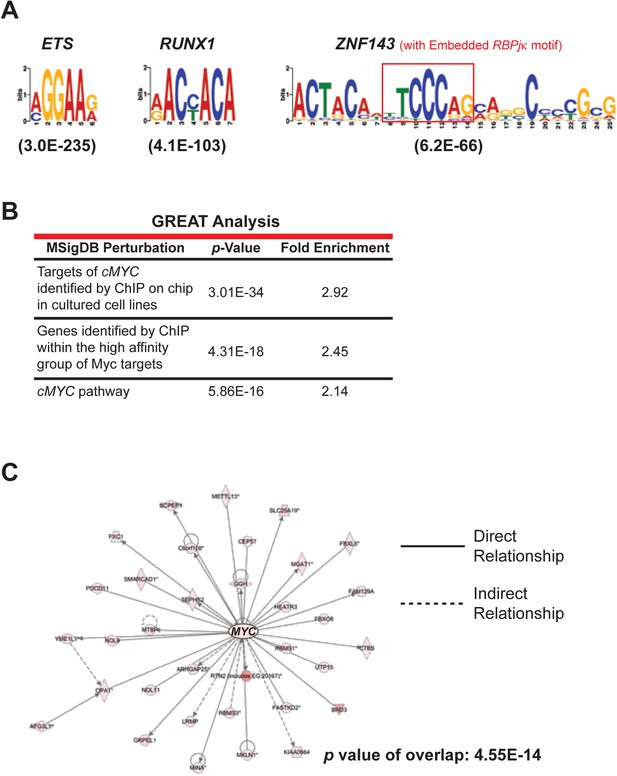
NOTCH ChIP-seq in ANGPTL2-stimulated CD34+ cells.
(A) Enriched transcription factor motifs at the peaks of NOTCH binding sites from NOTCH ChIP-seq. Parentheses indicate E values for the significance of the motif. An embedded RBPjκ ((consensus motif (red box) was found within the ZNF143 motif as previously described (Wang et al., 2011). (B) Examples of MYC-related categories from GREAT analysis of NOTCH ChIP-seq. (C) IPA network analysis of NOTCH ChIP-seq bound genes revealed a dense network of target genes centered on MYC.
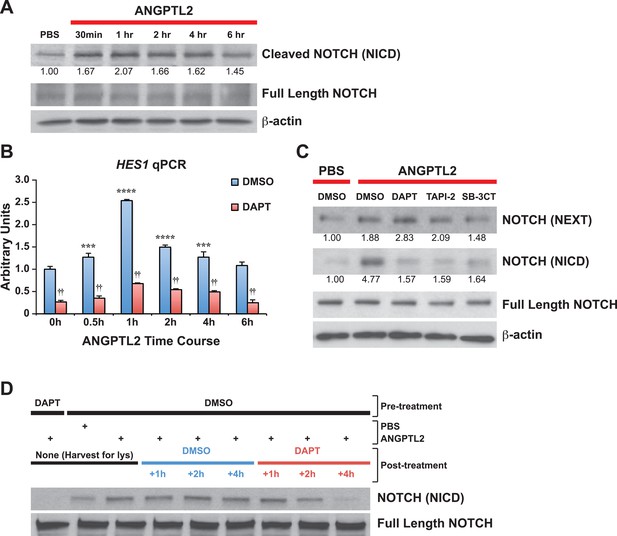
ANGPTL2 activates NOTCH by receptor cleavage.
(A) Human CD34+ cells were stimulated with ANGPTL2 (1 μg/ml) or PBS vehicle and Western blotted for S3 cleaved (NICD) and full-length NOTCH receptor. β-actin was used as loading control. Ratios indicated below the Western blot represent densitometry of the band intensity normalized to loading control. (B) Human endothelial cells pre-treated for 4 hr with DMSO vehicle or DAPT (20 μM) assayed for HES1 by qPCR. Error bars denote S.E.M., p < 0.0001, two way ANOVA; ***p < 0.001, ****p < 0.0001 compared to 0 hr and ††p < 0.0001 compared to DMSO, Bonferroni post-hoc test. (C) Human endothelial cells pre-treated with DMSO, DAPT (20 μM), TAPI-2 (20 μM), or SB-3CT (30 μM) prior to stimulating with ANGPTL2 (1 μg/ml) or PBS (1 hr) and Western blotted for S2 cleaved (NEXT), S3 cleaved (NICD), or full-length NOTCH. β-actin was used as loading control. Ratios indicated below the Western blots represent densitometry of the band intensity normalized to loading control. (D) NICD degradation experiment: post-treatment of ANGPTL2-stimulated cells (1 μg/ml for 1 hr) with DMSO or DAPT for 1, 2, or 4 hr to prevent de novo formation of NICD product, assayed by Western blotting. All experiments were repeated at least three times.
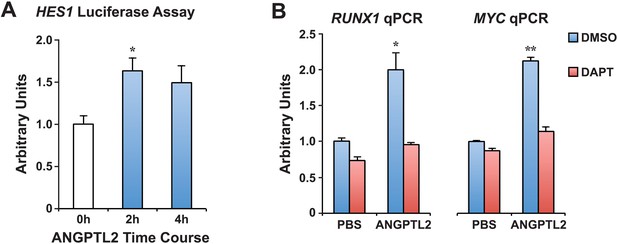
ANGPTL2 activates NOTCH signaling.
(A) K562 cells assayed for HES1 transcriptional activity by measuring the transfected HES1-responsive firefly luciferase activity normalized to the co-transfected internal control of Renilla luciferase activity after ANGPTL2 (1 μg/ml) stimulation. Error bars denote S.E.M., *p < 0.05 compared to 0 hr (white bar), Student's t test. (B) Similar qPCR performed as in Figure 5B for RUNX1 or MYC mRNA. Error bars denote S.E.M., *p < 0.05, **p < 0.01 compared to PBS DMSO vehicle control, Student's t test.
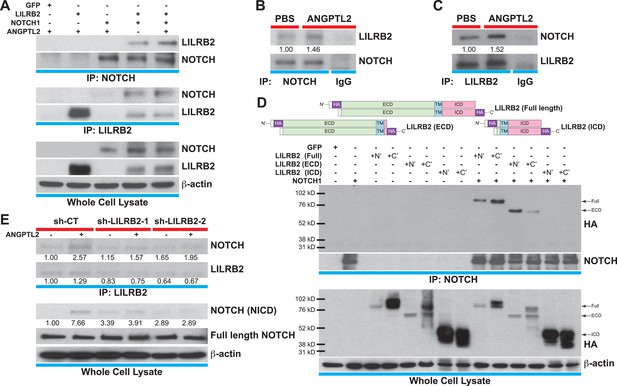
ANGPTL2 receptor, LILRB2, interacts with NOTCH in cis.
(A) Cell lysates from ANGPTL2-stimulated HEK293T cells transfected with GFP control, full-length LILRB2 and/or NOTCH1 plasmids were IP for NOTCH1 or LILRB2. Note the interaction between both receptors and this association increased upon ANGPTL2 (1 μg/ml, 1 hr) stimulation. Whole cell lysates were used for loading control. (B and C) Cell lysates from human CD34+ cells were co-IP for endogenous NOTCH1, endogenous LILRB2 or IgG isotype and Western blotted for associated LILRB2 or NOTCH1, respectively. (D) Cell lysates from NOTCH1 and HA-tagged LILRB2 truncation mutant (schematics) transfected cells were co-IP for NOTCH1 and Western blotted for HA. NOTCH1 interaction was only observed with full-length LILRB2 or LILRB2-ECD. (E) Stable lentiviral knockdown of LILRB2 using two different sequences. Partial knockdown of LILRB2 was observed compared to the control scrambled shRNA (sh-CT). The interaction between LILRB2 and NOTCH1 was also decreased (top panels). The ANGPTL2-induced NOTCH receptor cleavage in sh-CT cells was abolished in sh-LILRB2 cells. Whole cell lysates were done for loading control. All experiments were repeated at least 3 times. Ratios indicated below the Western blots represent densitometry of the band intensity.
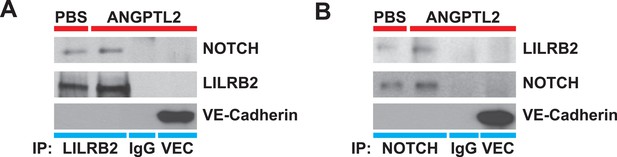
NOTCH interacts with LILRB2 in endothelial cells.
Angptl2 ANGPTL2-stimulated (1 μg/ml) human endothelial cell lysates were immunoprecipitated for (A) LILRB2, (B) NOTCH, IgG isotype, or VE-Cadherin (VEC, a highly abundant surface transmembrane protein) and Western blotted for NOTCH, LILRB2, and VEC to demonstrate antibody specificity for co-IP. Experiment repeated at least three times.
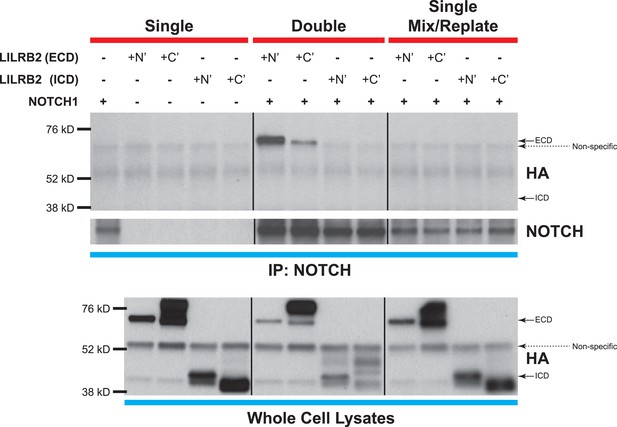
NOTCH and LILRB2 interact in cis.
Similar experiments as those performed in Figure 6D except cells were either singly transfected (left and right lanes) or doubly transfected (middle lanes) with LILRB2 ECD, LILRB2 ICD, and/or NOTCH1. Those that are singly transfected were then trypsinized before mixing and replating to exclude co-expression of LILRB2 and NOTCH within a single cell (right lanes). Cell lysates were co-IP for NOTCH and Western blotted for HA. Only the doubly transfected LILRB2 ECD and NOTCH showed interaction between the two (middle lanes). Whole cell lysates were used as loading controls. All experiments repeated at least 3 times.
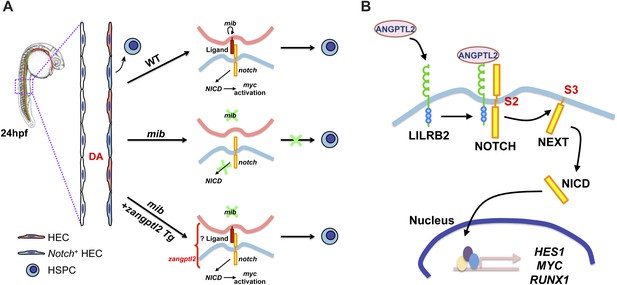
Proposed models for ANGPTL-mediated NOTCH signaling.
(A) Notch signaling in zebrafish is believed to be important for HSPC specification in the hemogenic endothelium on the ventral side of the developing dorsal aorta (DA). A simplified view of canonical notch signaling starts with mib endocytic processing of notch ligands to potentiate their ability to activate notch, depicted in the wild-type (WT) model. Notch receptors interact with ligands in the neighboring cell and this leads to subsequent cleavage of notch to release NICD to translocate into the nucleus and initiate transcription of target gene like myc. In the mib mutant however, the lack of signal from notch ligands prevents further downstream signaling from the receptor and HSPCs are not formed. From our genetic interaction studies, we believe that zangptl2 is downstream of mib (Figure 2C) but upstream of NICD generation (Figure 2B). (B) Using cultured cells to further explore the mechanism by which ANGPTL2 can regulate NOTCH signaling, we found that ANGPTL2 can stimulate NOTCH receptor cleavage at S2 and S3, leading to transcription of target genes. This was dependent on the ANGPTL receptor, LILRB2, as ANGPTL2 can induce recruitment of LILRB2 to NOTCH.
Videos
Time-lapse video of AGM budding in embryos transiently expressing constitutively active NOTCH.
Representative video depicting the HSPCs budding from the AGM in Tg(drl:eGFP;kdrl:NICD)-injected Tg(kdrl:Hras-mCherry);casper. Hemogenic endothelial cells (red) lining the ventral wall of the dorsal aorta that express the injected transgene (green) in the top panel depict at least three cell budding events (numbered with white arrow in the bottom panel showing GFP alone). Time-lapse imaging was captured at 10 min/frame and rendered at 3 frames/s. Each frame is a maximum projection of confocal z-stack, cropped from the entire AGM. Scale bar: 25 μm. DA: dorsal aorta; PCV: posterior cardinal vein.
Time-lapse video of AGM budding in control injected embryo.
Representative video depicting the DA of control Tg(drl:eGFP;kdrl:eGFP) injected Tg(kdrl:Hras-mCherry);casper. In this example, the two cells that contain the control transgene never budded off from the AGM during the 24 hr imaged. Videos were captured on the same day as NICD-injected embryos.
Additional files
-
Supplementary file 1
List of gene sets with significant positive enrichment score from Gene Set Enrichment Analysis (GSEA) of ANGPTL2-stimulated human CD34+ cells expression data. ANGPTL2 microarray was compared to 5562 gene sets in the Broad MSigDB (c2 curated gene sets, c3 motif gene sets, and c5 GO gene sets). 2521 gene sets showed a positive correlation (i.e., upregulated in ANGPTL2-stimulated cells). 284 gene sets were highly significant with FDR q-value of <0.05 and are highlighted in yellow. Those highlighted in red are Myc-related.
- https://doi.org/10.7554/eLife.05544.019





