Viral-inducible Argonaute18 confers broad-spectrum virus resistance in rice by sequestering a host microRNA
Figures
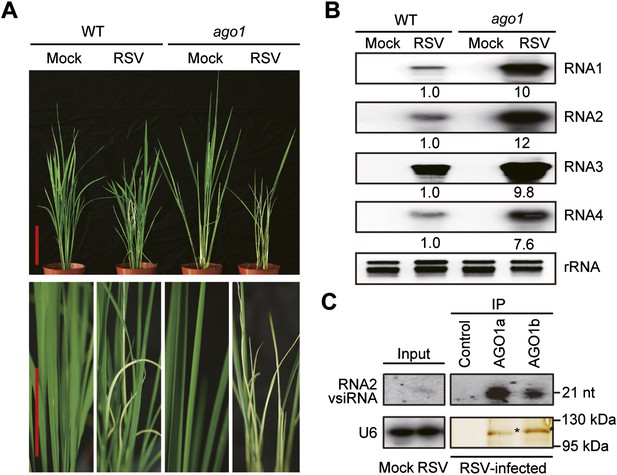
AGO1 participates in antiviral immunity in rice.
(A) Symptoms of WT and ago1 RNAi rice plants infected with RSV, pictures were taken at 6 weeks post-inoculation. Scale bars, 15 cm. (B) Detection of RSV genomic RNA segments in WT and ago1 RNAi rice plants by Northern blot. The blots were hybridized with radiolabeled riboprobes specific for each RNA segment. rRNAs were stained with ethidium bromide and served as loading controls. The RNA signals were quantified and normalized to rRNAs, and the relative values were calculated by comparison with those in RSV-infected WT (arbitrarily set to 1.0). (C) Detection of vsiRNAs associated with AGO1a and AGO1b immunoprecipitates. The silver-stained gel shows that comparable amounts of AGO1 complexes were used for RNA preparation. The asterisks indicate the positions of AGO proteins. The position of a RNA size marker, electrophoresed in parallel, is shown to the right of the blots. U6 was also probed and served as a loading control.
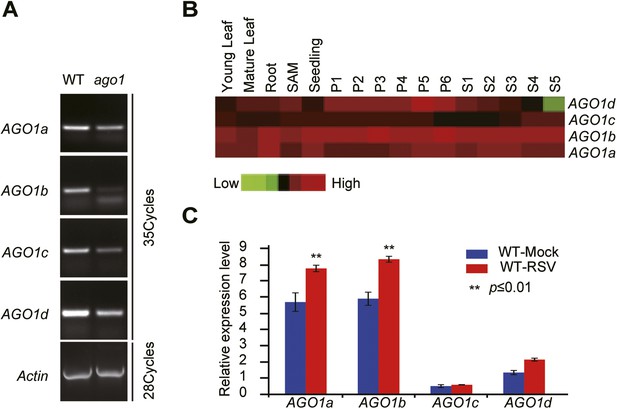
Characterization of rice ago1 RNAi lines and expression profiles of rice AGO1a, 1b, 1c, and 1d.
(A) Detection of AGO1a, AGO1b, AGO1c, and AGO1d in WT and the ago1 RNAi line by semi-quantitative RT-PCR. Actin was detected and used as a control. (B) Expression profiles of rice AGO1a, 1b, 1c, and 1d in vegetative tissues (young leaf, mature leaf, and root), shoot apical meristem (SAM), panicle at six different developmental stages (P1–P6), and seeds at five different developmental stages of seed development (S1–S5) examined by microarray analysis. (C) Detection of AGO1a, AGO1b, AGO1c, and AGO1d in mock- or RSV-inoculated rice plants by qRT-PCR. The expression levels were normalized using the signal from OsEF-1a. The average (± standard deviation) values from three biological repeats of qRT-PCR are shown.
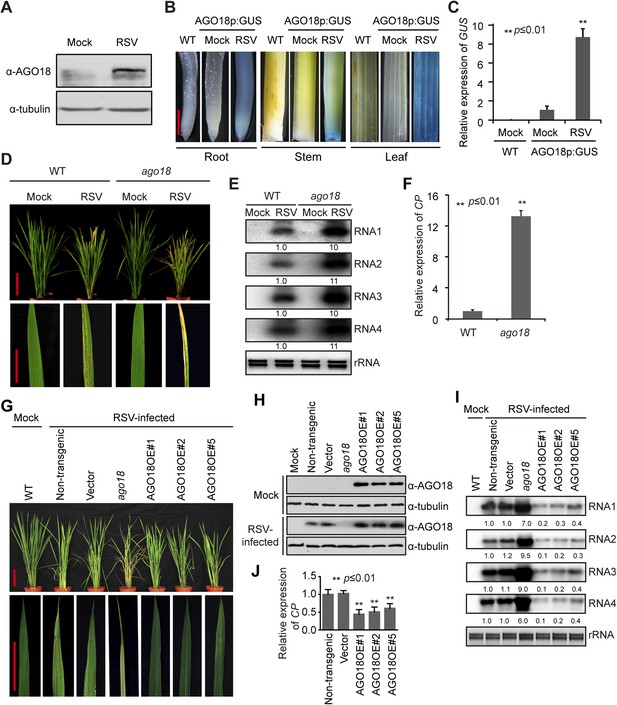
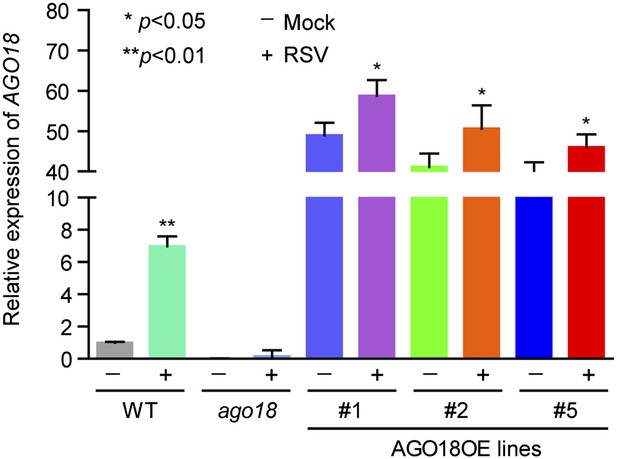
AGO18 is induced by viral infection and confers antiviral immunity in rice.
(A) Detection of AGO18 in mock- or RSV-inoculated rice plants by Western blot. Tubulin was probed and served as a loading control. (B) Representative GUS staining images of mock- or RSV-inoculated AGO18p:GUS transgenic plants. Scale bars, 5 mm. (C) qRT-PCR analysis of the transcript level of GUS in the indicated plants. The average (± standard deviation) values from three biological repeats of qRT-PCR are shown. (D) Symptoms of wild-type (WT) and ago18 rice plants infected with RSV, pictures were taken at 6 weeks post-inoculation. Scale bars, 15 cm. (E) Detection of RSV genomic RNA segments in WT and ago18 rice plants by Northern blot. The blots were hybridized with radiolabeled riboprobes specific for each RNA segment. rRNAs were stained with ethidium bromide and served as loading controls. The RNA signals were quantified and normalized to rRNAs, and the relative values were calculated by comparison with those in RSV-infected WT (arbitrarily set to 1.0). (F) Detection of RSV CP in the RSV-infected WT and ago18 by qRT-PCR. The expression levels were normalized using the signal from OsEF-1a. The average (± standard deviation) values from three biological repeats of qRT-PCR are shown. (G) Symptoms of RSV-infected WT (Non-transgenic), ago18 and transgenic lines overexpressing AGO18, pictures were taken at 6 weeks post-inoculation. Scale bars, 15 cm (upper panel) and 5 cm (lower panel). (H) Detection of AGO18 in mock- or RSV-inoculated plants as indicated. Tubulin was probed and served as a loading control. (I) Detection of RSV genomic RNA segments in the indicated plants by Northern blot. The blots were hybridized with radiolabeled riboprobes specific for each RNA segment. rRNAs were stained with ethidium bromide and served as loading controls. The RNA signals were quantified and normalized to rRNAs, and the relative values were calculated by comparison with those in RSV-infected WT (Non-transgenic) (arbitrarily set to 1.0). (J) Detection of RSV CP in the indicated RSV-infected plants by qRT-PCR. The expression levels were normalized using the signal from OsEF-1a. The average (± standard deviation) values from three biological repeats of qRT-PCR are shown.
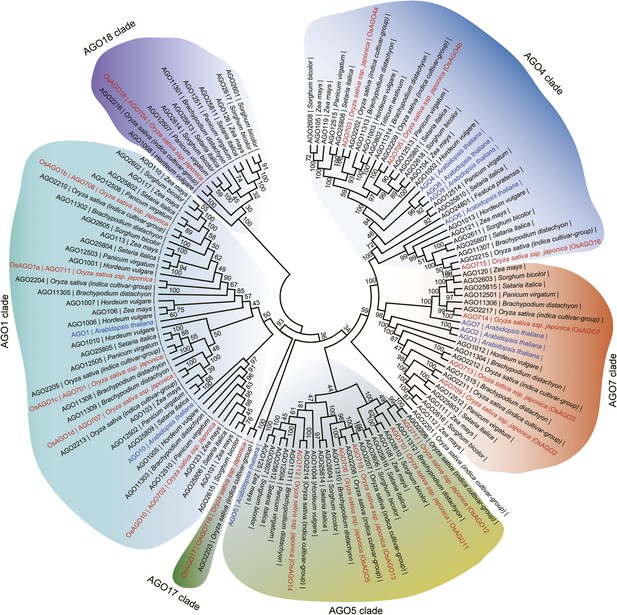
Phylogenetic analysis of AGO family proteins in plants.
Unrooted, neighbor-joining tree was constructed by alignments of AGO protein sequences from the indicated plants.
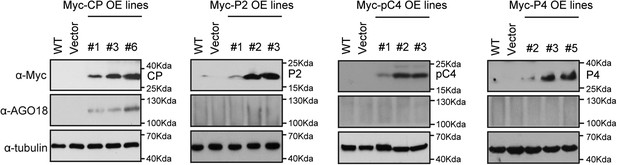
RSV CP triggers AGO18 expression.
Detection of CP, P2, pC4, P4, and AGO18 in transgenic rice lines that overexpress RSV CP, P2, pC4, and P4 by Western blot analysis. Tubulin was also detected and served as a loading control.
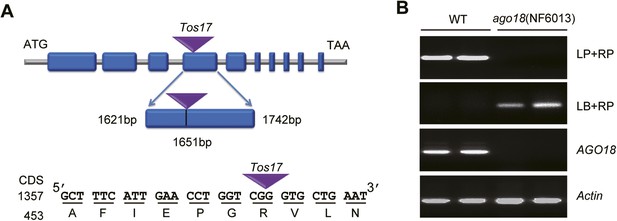
Identification of the ago18 mutant.
(A) A diagram showing the site of Tos17 insertion in AGO18 locus in the ago18 mutant. (B) Upper two panels: Genotyping of the ago18 mutant by PCR. LP: left primer; RP: right primer; LB: left T-DNA (Tos17) border primer. Lower two panels: RT-PCR analysis of AGO18 expression in wild-type (WT) and ago18 plants. Actin was detected and served as a control.
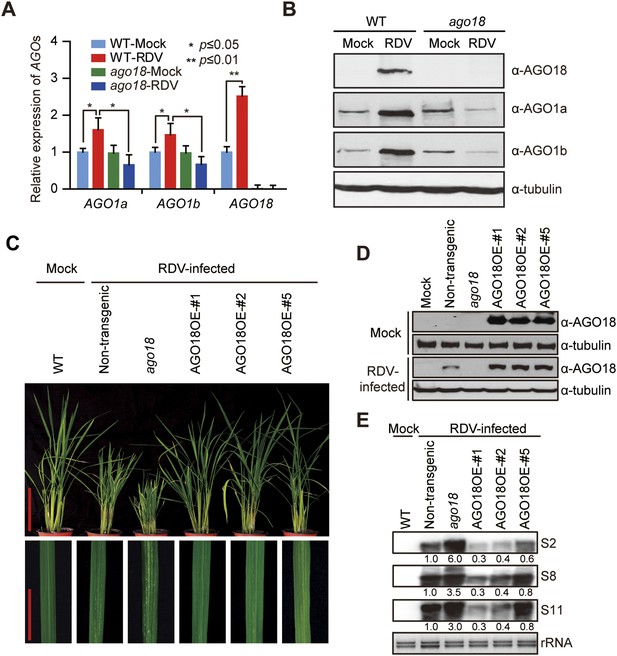
AGO18 is required for rice resistance to RDV infection.
(A) qRT-PCR analysis of AGO1 and AGO18 expression levels in mock- or RDV-inoculated WT and ago18 plants. The expression levels were normalized using the signal from OsEF-1a. The average (± standard deviation) values from three biological repeats of qRT-PCR are shown. (B) Detection of AGO1a, AGO1b, and AGO18 proteins in mock- or RDV-inoculated WT and ago18 plants. Tubulin was detected and served as a loading control. (C) Symptoms of RDV-infected WT (Non-trangenic), ago18 and transgenic lines overexpressing AGO18, pictures were taken at 6 weeks post-inoculation. Scale bars, 15 cm (upper panel) and 5 cm (lower panel). (D) Detection of AGO18 in mock- or RDV-inoculated plants as indicated. Tubulin was probed and served as a loading control. (E) Detection of RDV genomic RNA segments in the indicated plants by Northern blot. The blots were hybridized with radiolabeled riboprobes specific for each RNA segment. rRNAs were stained with ethidium bromide and served as loading controls. The RNA signals were quantified and normalized to rRNAs, and the relative values were calculated by comparison with those in RSV-infected WT (arbitrarily set to 1.0).
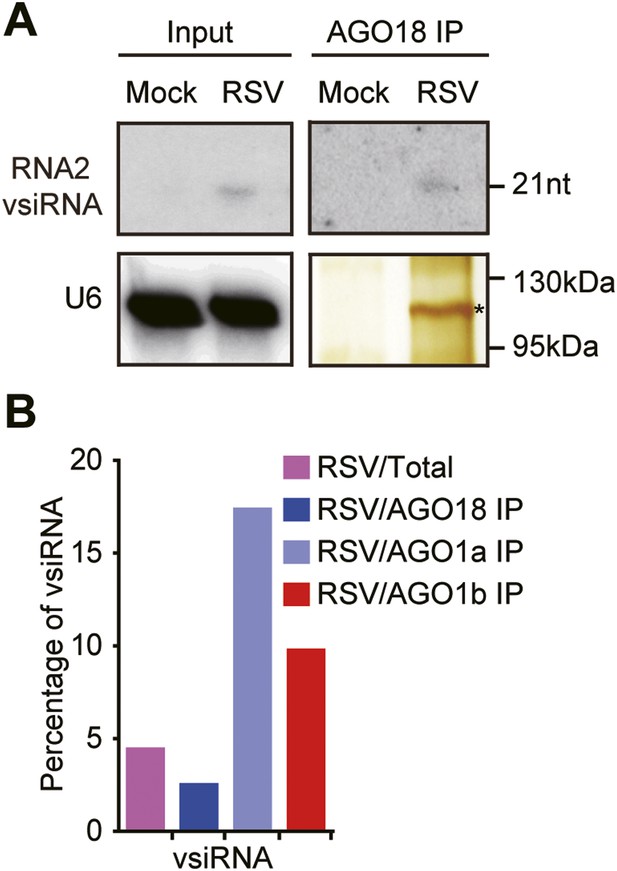
AGO18 unlikely functions as an effector of vsiRNAs.
(A) Detection of vsiRNAs in total extracts (Input) or AGO18 immunoprecipitates prepared from mock- or RSV-inoculated plants by Northern blot. The silver-stained gel shows the quality of purified AGO18 complexes. The asterisk indicates the position of AGO18. The position of an RNA size marker is shown on the right of the blot. U6 is probed and served as a loading control. (B) Percentage of deep sequencing reads matching vsiRNAs in total reads obtained from total extracts, AGO1a, AGO1b, and AGO18-associated small RNAs. Samples for deep sequencing were prepared from RSV-infected rice plants.
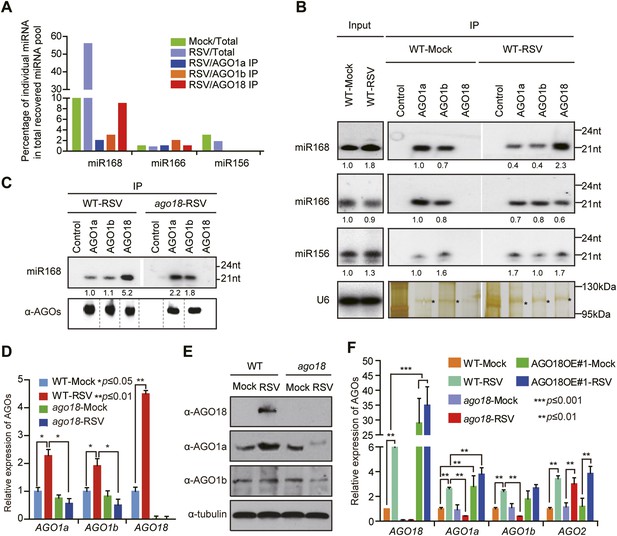
AGO18 competes with AGO1 for miR168 to up-regulate AGO1 upon viral infection.
(A) Percentage of deep sequencing reads matching the indicated miRNAs in total reads obtained from total extracts, AGO1a, AGO1b, and AGO18-associated small RNAs. Samples for deep sequencing were prepared from mock- or RSV-inoculated rice plants. (B) Detection of the indicated miRNAs in total extract (Input), AGO1a, AGO1b, and AGO18 complexes by Northern blot. The blots were stripped and reprobed for multiple times. The silver-stained gel shows that comparable amounts of different AGO complexes were used for RNA preparation. The asterisks indicate the positions of AGO proteins. The positions of RNA size markers are shown on the right of the blots. The RNA signals were quantified, and the relative values were calculated by comparison with those in total extracts or AGO1a complex prepared from mock-inoculated WT (arbitrarily set to 1.0). (C) Northern blot analysis showing miR168 from AGO1a, AGO1b, and AGO18 complexes in RSV-infected WT and ago18 plants (upper panel). Western blot gel shows that comparable amounts of different AGO complexes were used for RNA preparation (lower panel). (D) qRT-PCR analysis of the levels of AGO1a, AGO1b, and AGO18 in WT rice and ago18 mutants with or without RSV infection. The expression levels were normalized using the signal from OsEF-1a. The average (± standard deviation) values from three biological repeats of qRT-PCR are shown. (E) Western blot showing AGO1a, AGO1b, and AGO18 protein levels in WT and ago18 with or without RSV infection. Tubulin was probed and served as a loading control. (F) qRT-PCR analysis of the levels of AGO1a, AGO1b, AGO2, and AGO18 in the indicated plants. The expression levels were normalized using the signal from OsEF-1a. The average (± standard deviation) values from three biological repeats of qRT-PCR are shown.
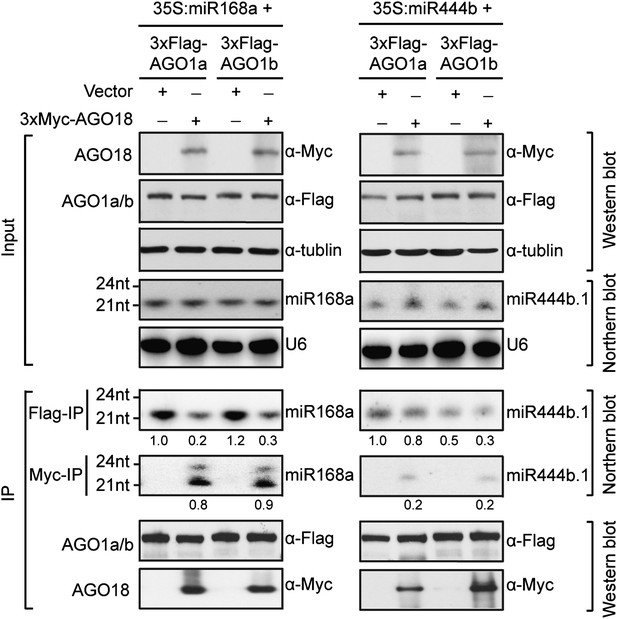
AGO18 competes with AGO1 for miR168 in vitro.
Specific AGO18–miR168 interaction was confirmed by in vitro assays in N. benthamiana. Constructs with the indicated combinations were introduced into N. benthamiana leaves for transient expression by agro-infiltration. Northern blots were conducted with total RNA (Input) and small RNAs recovered from immunoprecipitated AGO complexes (IP). Western blot analyses were done with the crude extract and aliquots of the IP products using anti-Flag or anti-Myc antibodies. The positions of RNA size markers are shown on the left of the blots. U6 was probed and served as a loading control.
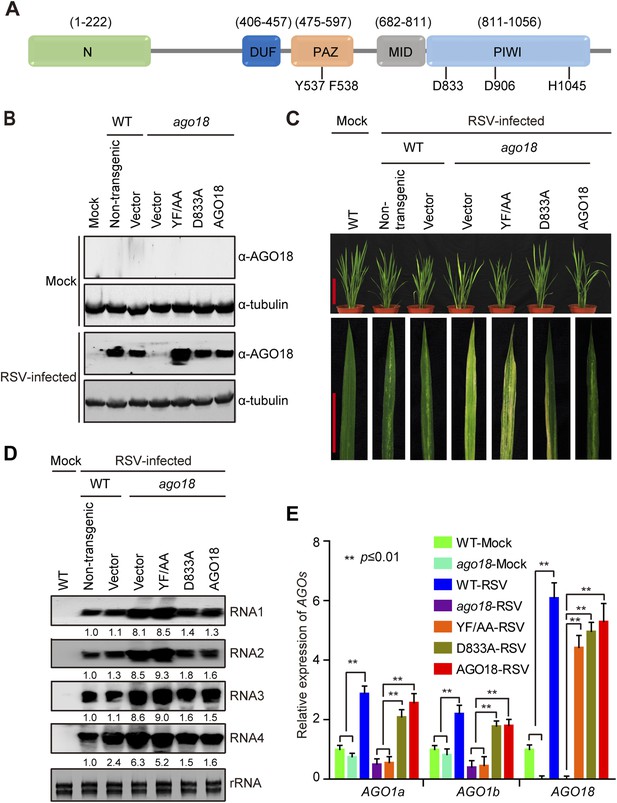
Small RNA-binding but not slicing activity of AGO18 is required for its antiviral function.
(A) Domain structure of AGO18 protein. AGO18 consists of a variable N-terminal domain and conserved C-terminal PAZ, MID, and PIWI domains. The residues Y537 and F538 required for small RNA binding, and D833, D906, and H1045 required for slicing are indicated. (B) Detection of AGO18 protein in mock- or RSV-inoculated WT (Non-trangenic) plants as well as ago18 mutants complemented with AGO18 and its derivatives by Western blot. Tubulin was probed and served as a loading control. (C) Symptoms of mock- or RSV-inoculated WT (Non-trangenic) plants as well as ago18 mutants complemented with AGO18 or its derivatives, pictures were taken at 6 weeks post-inoculation. Scale bars, 15 cm (upper panel) and 5 cm (lower panel). (D) Detection of RSV genomic RNA segments in mock- or RSV-inoculated WT (Non-trangenic) plants as well as ago18 mutants complemented with AGO18 or its derivatives by Northern blot. The blots were hybridized with radiolabeled riboprobes specific for each RNA segment. rRNAs were stained with ethidium bromide and served as loading controls. The RNA signals were quantified and normalized to rRNAs, and the relative values were calculated by comparison with those in RSV-infected WT (arbitrarily set to 1.0). (E) qRT-PCR analysis of the levels of AGO1a, AGO1b, and AGO18 in RSV-infected WT plants as well as ago18 mutants complemented with AGO18 or its derivatives. The expression levels were normalized using the signal from OsEF-1a. The average (± standard deviation) values from three biological repeats of qRT-PCR are shown.
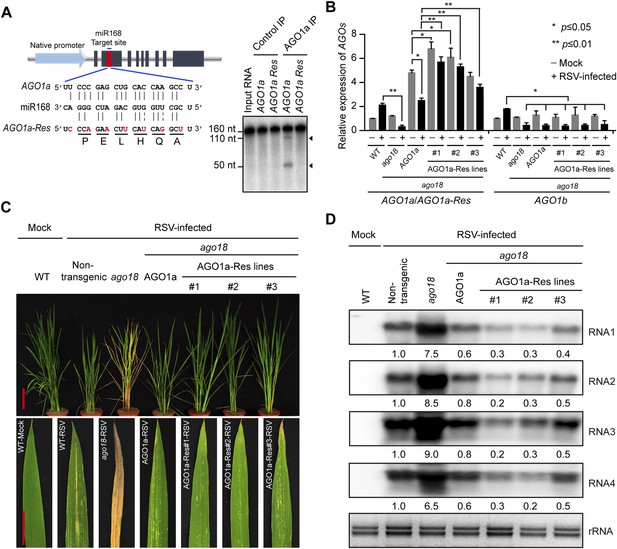
Transgenic expression of miR168-resistant AGO1a rescued the deficiency of ago18 for viral resistance.
(A) Schematic drawing of AGO1a featuring the target site of miR168. In AGO1a that is resistant to miR168 cleavage (AGO1a-Res), seven synonymous nucleotide substitutions were introduced into the miR168 target site. The resistance of AGO1a-Res to miR168-directed cleavage was examined by in vitro cleavage assay using purified AGO1a complex. Positions of the cleavage products are indicated by arrows. (B) qRT-PCR analysis of AGO1a and AGO1b expression levels in mock (−) or RSV-inoculated (+) WT, ago18, and transgenic plants expressing AGO1a or AGO1a-Res in the ago18 mutant background. The expression levels were normalized using the signal from OsEF-1a. The average (± standard deviation) values from three biological repeats of qRT-PCR are shown. (C) Symptoms of mock- or RSV-inoculated WT (Non-trangenic), ago18, and transgenic plants expressing AGO1a or AGO1a-Res in the ago18 mutant background. Scale bars, 15 cm. (D) Detection of RSV genomic RNA segments in the indicated plants by Northern blot. The blots were hybridized with radiolabeled riboprobes specific for each RNA segment. rRNAs were stained with ethidium bromide and served as loading controls. The RNA signals were quantified and normalized to rRNAs, and the relative values were calculated by comparison with those in RSV-infected WT (Non-trangenic) (arbitrarily set to 1.0).
Additional files
-
Supplementary file 1
(A) Disease incidence of RSV- or RDV-inoculated rice plants. (B) Summary of small RNA deep sequencing datasets. (C) Analysis of miRNA abundance by deep sequencing. (D) Oligonucleotides used in this study.
- https://doi.org/10.7554/eLife.05733.015
-
Supplementary file 2
Perl script used for statistical analysis of miRNA expression.
- https://doi.org/10.7554/eLife.05733.016