Autoinhibition of Bruton's tyrosine kinase (Btk) and activation by soluble inositol hexakisphosphate
Figures
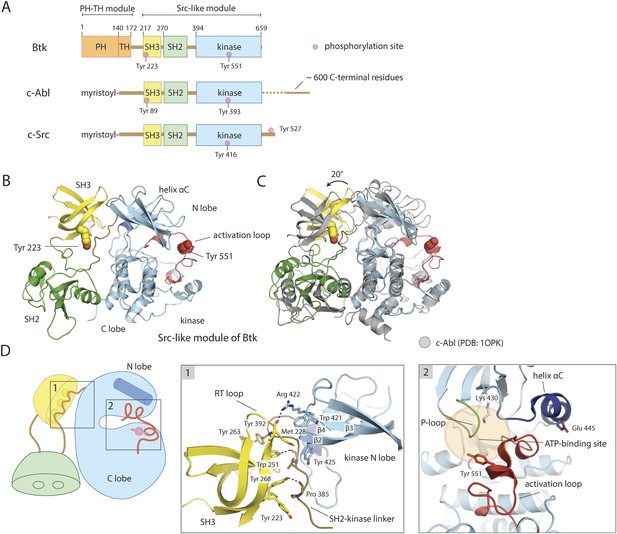
Crystal structure of the Src-like module of Btk.
(A) Domain architectures of Btk, c-Abl, and c-Src. (B) Model for the Src-like module of Btk, based on the crystal structure of the domain-swapped dimer. (C) Comparison of the Src-like modules of Btk and c-Abl (PDB: 1OPK) (Nagar et al., 2003), superimposed on the C lobes of the kinase domains. There is a relative rotation of about 15° for the SH2 domains and 20° for the SH3 domains for the two structures. (D) Details of the SH3/SH2-kinase linker interface and the kinase catalytic cleft in Btk. Left panel: a cluster of hydrophobic residues on the SH3 domain packs against the polyproline type-II helix formed by the SH2-kinase linker. This type of interaction is seen in most SH3-peptide ligand complexes. Right panel: the activation loop of Btk folds into the mouth of the catalytic cleft, blocking part of the ATP-binding cleft. Glu 445, in helix αC, forms an ion pair with Lys 430 in the active conformation but is prevented from doing so in this inactive conformation by the activation loop and helix αC.
-
Figure 1—source data 1
Structures of fragments of Tec family kinases in the Protein Data Bank.
- https://doi.org/10.7554/eLife.06074.017
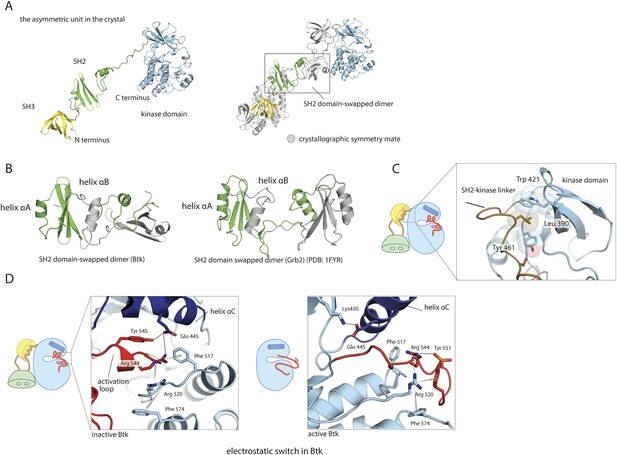
Structural details of the Src-like module of Btk.
(A) The Src-like module of Btk forms a domain-swapped dimer in the crystal lattice, with one molecule per asymmetric unit. The SH3 domain and the kinase domain are intact, but the SH2 domain forms a domain-swapped dimer with another molecule. (B) Molecular details of the domain-swapped SH2 dimer in Btk and Grb2 (Schiering et al., 2000). The SH2 domain opens up at a position within the β-sheet, so that helix αB of one polypeptide chain packs against the β-sheet of the other. (C) Interactions between the SH2-kinase linker and the kinase domain. Leu 390 of the linker stabilizes the inactive conformation of the kinase domain by being sandwiched between residues Trp 421 and Tyr 461 in the kinase domain. (D) The ‘electrostatic switch’ in the Btk kinase domain. The active conformation of the Btk kinase domain is modeled from the crystal structure of Lck (PDB: 3LCK) (Yamaguchi and Hendrickson, 1996) using Modeller (Eswar et al., 2006). Activation entails a shuffling of salt-bridge and polar hydrogen-bond partners.
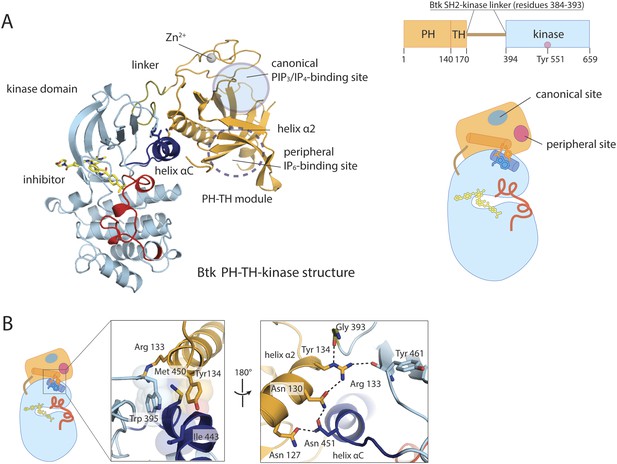
Crystal structure of the PH-TH-kinase construct of Btk.
(A) Crystal structure of the PH-TH-kinase construct. The PH-TH module is connected to the kinase domain via a 13-residue linker from the SH2-kinase linker of Btk. The kinase domain is bound to the inhibitor CGI1746 (Di Paolo et al., 2011). The canonical PIP3/IP4 binding pocket of the PH domain is located distal to the interface with the kinase, and the peripheral binding site for IP6 is indicated. (B) The interface between the PH-TH module and the kinase domain of Btk.
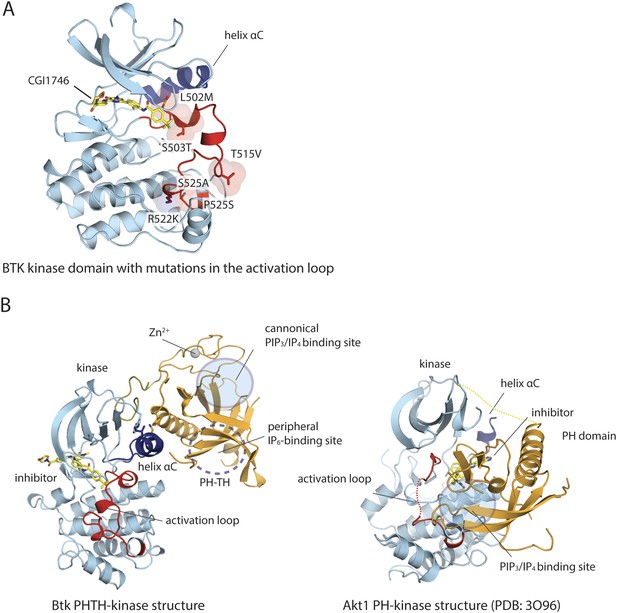
Structure of the kinase domain of Btk and its interaction with the PH domain.
(A) Crystal structure of the isolated Btk kinase domain with mutations in the activation loop. These mutations (L542M, S543T, V555T, R562K, S564A, and P565S) are based on a previous study (Joseph et al., 2013), and they improved the quality of the crystals of the PH-TH-kinase construct. The structure of the mutant kinase is very similar to that of the wild-type Btk kinase domain (PDB: 3OCS). (B) Comparison of the Btk PH-TH-kinase structure (left) and the Akt1 PH-kinase structure (right). In Akt1, The PH domain is docked between the N lobe and the C lobe of the kinase domain, and it blocks substrate access to the catalytic cleft. The lipid-binding site on the PH domain is buried at the PH-kinase interface, and binding of PIP3/IP4 breaks the PH-kinase interface and activates Akt1 (Wu et al., 2010; Calleja et al., 2009).
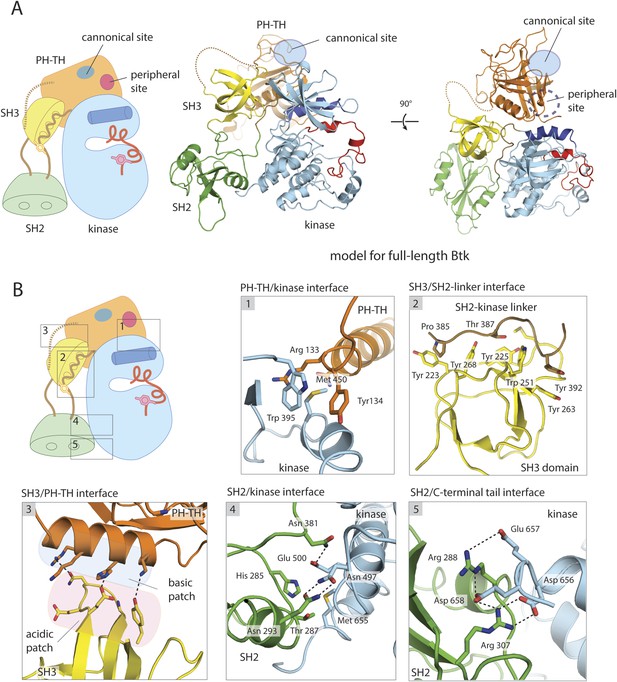
A structural model for full-length Btk.
(A) A composite model for full-length Btk. The PH-TH domain sits on top of the kinase domain and the SH3 domain, serving as a ‘latch’ that presumably stabilizes the Src-like module in the autoinhibited conformation. The 45-residue linker between the PH-TH module and the SH3 domain is represented by the dotted brown line. Two orthogonal views of an instantaneous structure at 360 ns from a molecular dynamics trajectory for the composite model are shown. (B) Inter-domain interactions in an instantaneous structure at 360 ns from a simulation of the composite model. The SH2/kinase interface and the PH-TH /SH3 interface are formed and remain stable during the molecular dynamics simulation.
-
Figure 3—source data 1
Structures from several time points in the simulation of the composite model for full-length Btk.
The structures are taken every 20 ns from a 360 ns molecule dynamic simulation of the composite model. See ‘Materials and methods’ for the details of simulations.
- https://doi.org/10.7554/eLife.06074.010
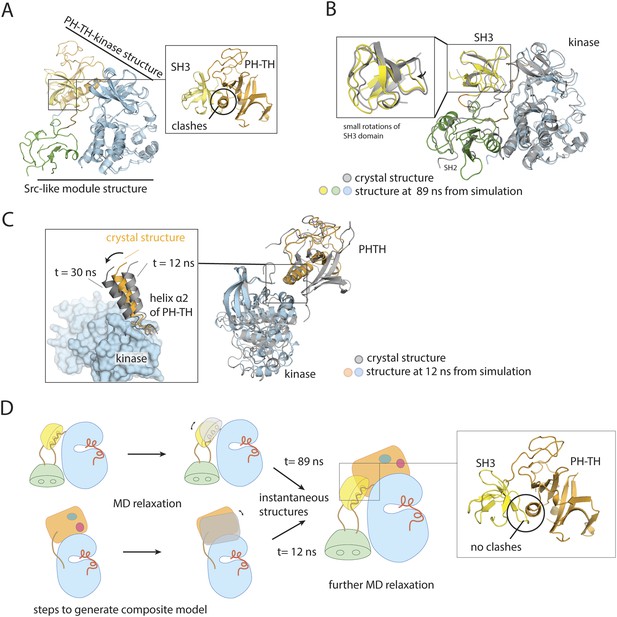
Utilization of molecular dynamics in constructing a model for full-length Btk.
(A) Overlay of the crystal structures of the Btk Src-like module and the PH-TH-kinase construct, using the kinase domain C lobe as the reference. Sidechain clashes (circled) are observed between helix α2 in the PH-TH module and the β3/β4 loop segment in the SH3 domain. (B) Fluctuations in the Btk Src-like module occurred during a 100 ns molecular dynamics simulation. An instantaneous structure (t = 89 ns) from the simulation is overlaid on the crystal structure of the Src-like module. (C) Fluctuations in the Btk PH-TH-kinase construct during a 100 ns molecular dynamics simulation. An instantaneous structure (t = 12 ns) from the simulation is overlaid on the crystal structure of the PH-TH-kinase construct using the kinase domain N lobe as the reference. The PH-TH module pivots (black arrow) about an apparent anchor point at the C terminus of helix α2. (D) Steps used to generate a composite model for full-length Btk. An instantaneous structure (t = 89 ns) of the Src-like module and an instantaneous structure (t = 12 ns) of the PH-TH-kinase construct from the molecular dynamics trajectory are overlaid, using the kinase domain C-lobe as the reference. In contrast to a corresponding overlay of the crystal structures, there are no sidechain clashes between helix α2 in the PH-TH module and the loop in the SH3 domain. A model for full-length Btk is obtained by combining the PH-TH module with the Src-like module in the two instantaneous structures, and is subject to further molecular dynamics simulation.
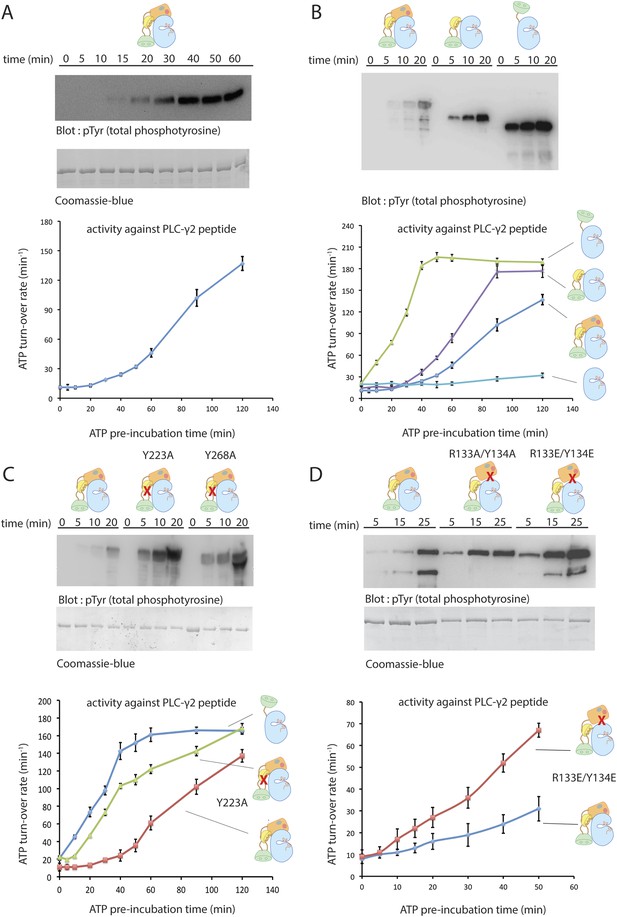
Autoinhibition of Btk.
(A) Activation of full-length bovine Btk (residues 1 to 659, 2 μM). Reactions are carried out in the presence of 10 mM Mg2+, 150 mM NaCl, 1 mM ATP, 25 mM Tris-HCl pH 8.0. The level of autophosphorylation is assayed by immunoblotting an SDS-PAGE gel with a non-specific, anti-phosphotyrosine antibody (4G10, EMD Millipore) (upper panel). The amount of total protein loaded on the gel is measured by coomassie-blue staining. The kinase activity of Btk is assayed by a continuous kinase-coupled colorimetric assay, in the presence of 1 mM PLC-γ2 peptide substrate. See methods for detailed experimental procedures. (B) Comparison of the activation of the Btk Src-like module (residues 217 to 659), SH2-kinase (residues 270 to 659), and the kinase domain (residues 394 to 659). The SH2-kinase construct activates substantially faster than full-length Btk and the Src-like module of Btk. Activated full-length Btk degrades to a small extent over time, which results in some lower molecule-weight bands being detected on the western blot. (C) Activation of full-length Btk with mutations Y223A and Y268A. Tyr 223 and Tyr 268 are on the SH3/SH2-linker interface, and the two mutants activate faster than wild-type Btk. (D) Activation of full-length Btk with a double mutation (R134E/Y133E). Arg 134 and Tyr 133 are located at the PH-TH/kinase interface.
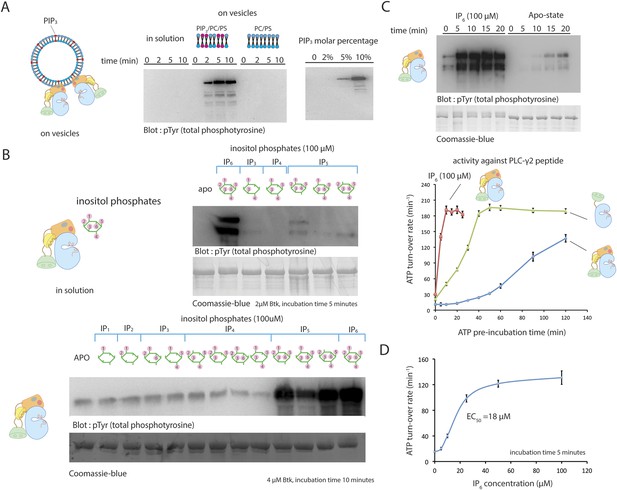
Activation of Btk.
(A) Activation of full-length Btk on membranes. Left panel: Btk (1 μM) autophosphorylates rapidly on lipid vesicles (total lipid concentration, 1 mM) containing 75% DOPC, 20% DOPS and 5% PIP3. Right panel: end-point autophosphorylation assay of Btk in the presence of lipid vesicles. The molar concentration of DOPC is kept at 75%, and the lipid fractions of PIP3 is 0%, 2%, 5%, and 10%, with the remaining lipid fraction filled with DOPS. The measurements are performed after 2 min incubation with lipid vesicles in the presence of ATP/Mg2+. (B) End-point autophosphorylation assays of Btk in solution in the presence of inositol phosphates. Upper panel: Btk (2 μM) autophosphorylates rapidly in the presence of 100 μM IP6 in solution, with the results after the first 5 min of incubation shown here. Essentially no phosphorylation is detected in Btk samples incubated with IP3, IP4 or I(1,3,4,5,6)P5, and very low phosphorylation is detected in Btk samples incubated with I(1,2,3,5,6)IP5 and I(2,3,4,5,6)IP5 in the same experiment. Lower panel: the assay is repeated with other inositol phosphates at a higher protein concentration (4 μM) and longer incubation time (10 min), and the gel was exposed for a longer time. There is a small amount of degraded Btk in the sample, as judged by the coomassie staining, leading to multiple bands in the western blot shown here and in panel C. (C) Activation of full-length Btk (2 μM) in the presence of IP6 (100 μM). (D) Activation of full-length Btk (2 μM) with increasing concentrations of IP6.
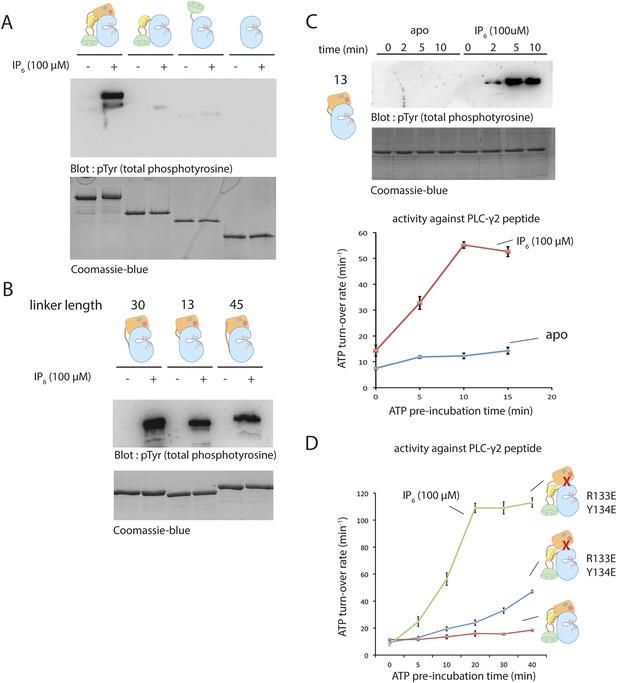
Activation of various deletion constructs of Btk.
(A) End-point autophosphorylation assay for full-length Btk, the Src-like module, the SH2-kinase construct and the kinase domain in the presence/absence of IP6. The measurements are carried out after incubating proteins (2 μM) with and without IP6 (100 μM) for 2 min in the presence of ATP/Mg2+. (B) End-point autophosphorylation assay for Btk PH-TH-kinase constructs of various linker lengths in the presence/absence of IP6 (100 μM). The measurements are carried out after incubating proteins (2 μM) with and without IP6 (100 μM) for 2 min at the presence of ATP/Mg2+. (C) Activation of the PH-TH-kinase construct (2 μM) with a 13-residue linker in the presence and absence of IP6 (100 μM). (D) Activation of full-length Btk (2 μM) with mutations R133E/Y134E in the presence and absence of IP6 (100 μM). Arg 133 and Tyr 134 are located at the PH-TH/kinase interface.
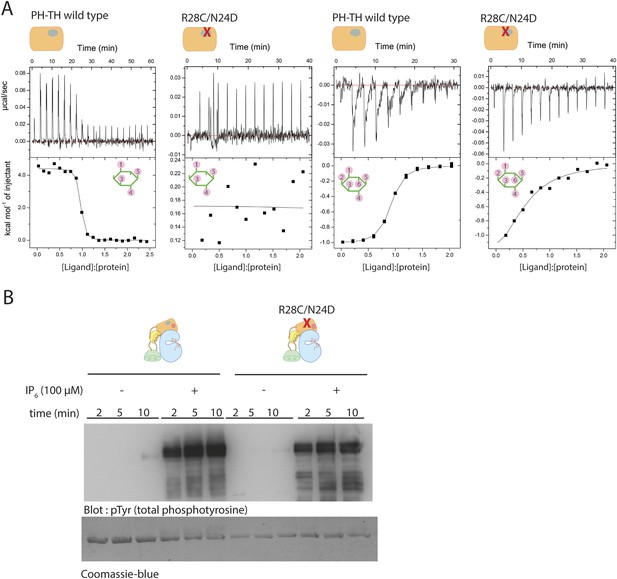
Binding of IP4 and IP6 to the Btk PH-TH module.
(A) Representative isothermal titration calorimetry data for the Btk PH-TH module and its R28C/N24D variant binding to IP4 or IP6. The protein concentration is 20 μM and that of the inositol phosphates goes up to 300 μM. Experiments are performed at 20°C. Titration curves are shown with the baseline-corrected raw data. The parameters from fitting a one-site binding model are listed in Table 3. (B) Activation of full-length Btk N24D/R28C mutant (2 μM) in the presence and absence of IP6 (100 μM). Asn 24 and Arg 28 are critical residues for PIP3/IP4 binding in the canonical lipid-binding pocket of the PH-TH module.
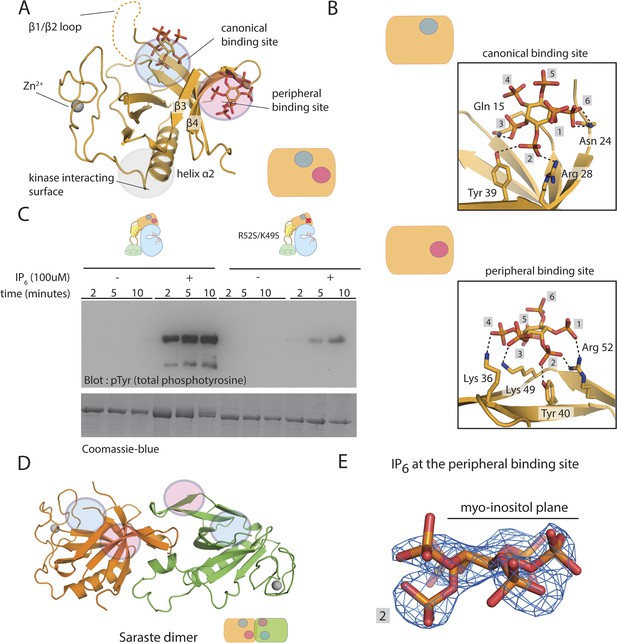
Crystal structure of the PH-TH module bound to IP6.
(A) Structure of the PH-TH module bound to two IP6 molecules. One IP6 molecule is in the canonical lipid-binding site and the other IP6 molecule is in the peripheral binding site formed by strands β3 and β4. The β1/β2 loop (dotted lines) is disordered in this structure. (B) IP6 coordination in the canonical binding site and in the peripheral binding site. (C) Activation of the Btk R52S/K49S mutant (2 μM) in the presence and absence of IP6 (100 μM). Arg 52 and Lys 49 are two residues interacting with IP6 in the peripheral binding site. (D) A dimer assembly of the Btk PH-TH module in the crystal structure of the IP6-bound PH-TH module. The canonical binding site and the peripheral binding site are shown in blue and magenta circle, respectively. We refer to this dimer as the ‘Saraste dimer’ (Hyvönen and Saraste, 1997). (E) An electron density map, using σA-weighted 2m|Fo| − D|Fc| coefficients (Read, 1986) for IP6 at the peripheral binding site, contoured at 1.5 σ.
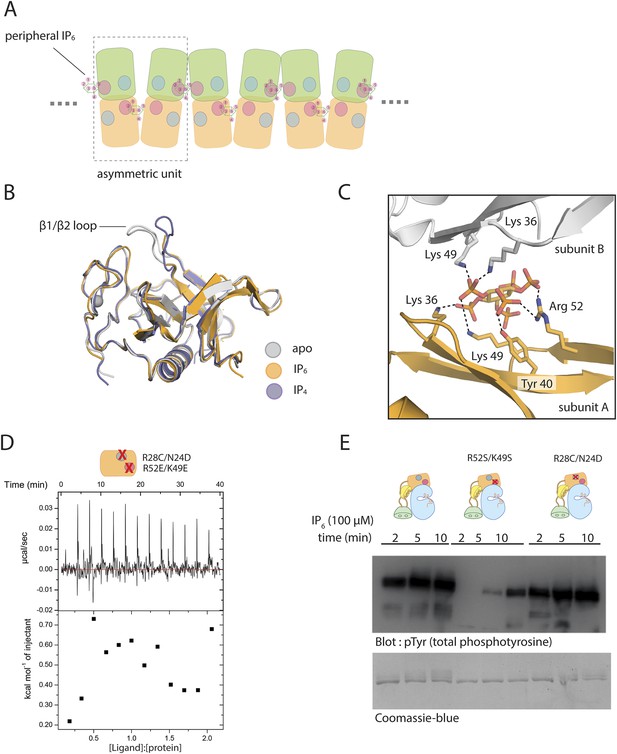
Aspects of the binding of IP6 to Btk.
(A) An open-ended chain of PH-TH dimers in the crystal lattice, mediated by IP6 at the pheripheral site. IP6 is bound at the peripheral site in such a way that only one IP6 molecule occupies the two sites at each interface. (B) Structure superposition of the IP6-bound PH-TH module with apo- and the IP4-bound PH-TH module. The β1/β2 loop, which coordinates IP4 binding, adopts a different conformation in the apo-structure, and is partially disordered in the IP6-bound structure. (C) IP6 coordination in the peripheral binding site by a crystal symmetry mate of the PH-TH module in the crystal structure of IP6-bound PH-TH module. IP6 and the residues involved in coordination are shown in sticks. The hydrogen bonds are shown in dotted lines. This is a detailed view of the interfacial sites shown in the schematic diagram in Panel A. (D) Isothermal titration calorimetry data for Btk PH-TH mutant R52E/K49E/R28C/N24D titrated with IP6. Arg 52 and Lys 49 are critical residues coordinating IP6 binding in the peripheral site. Arg 28 and Asn 24 are critical residues coordinating IP6 binding in the canonical site. Note that IP6 does not bind to this mutant form of the PH-TH module. (E) Activation of full-length Btk, Btk R52S/K49S mutant, and Btk R28C/N24D mutant in the presence of IP6 (100 μM).
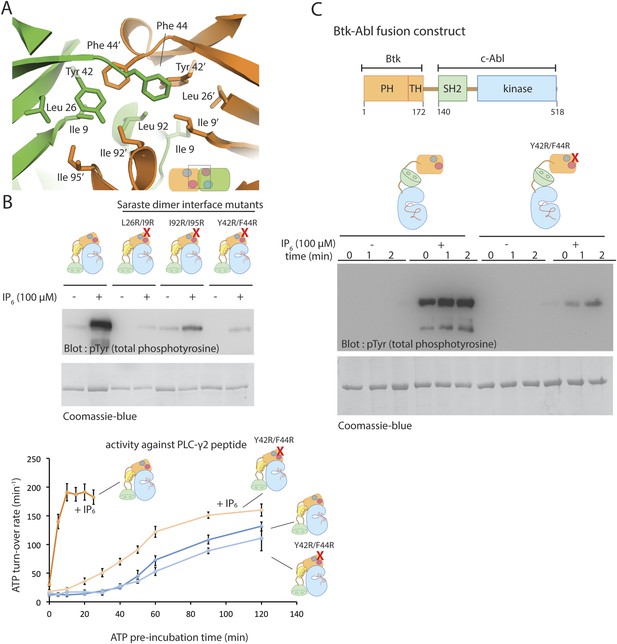
The Saraste dimer of the PH-TH module is critical for Btk activation by IP6.
(A) Molecular details of the Saraste dimer interface in the crystal structure of the IP6-bound PH-TH module. (B) End-point autophosphorylation assays for wild-type Btk, Btk L29R/I9R mutant, Btk I94R/I95R mutant, and Btk Y42R/F44R mutant in the presence and absence of IP6 (100 μM). The measurements were made after incubating the proteins with and without IP6 in the presence of ATP/Mg2+ for 5 min. Residues Leu 29, Ile 9, IIe 94, IIe 95 Tyr 42 and Phe 44 are located in the Saraste dimer interface, as shown in panel A. Activation of the Btk Y42R/F44R mutant is shown in the lower panel. (C) Activation of the Btk-Abl fusion construct (1 μM) and that of a variant of this fusion protein in which the Btk PH-TH module is mutated (Y42R/F44R). Measurements were made with 1 μM protein in the presence and absence of IP6 (100 μM).
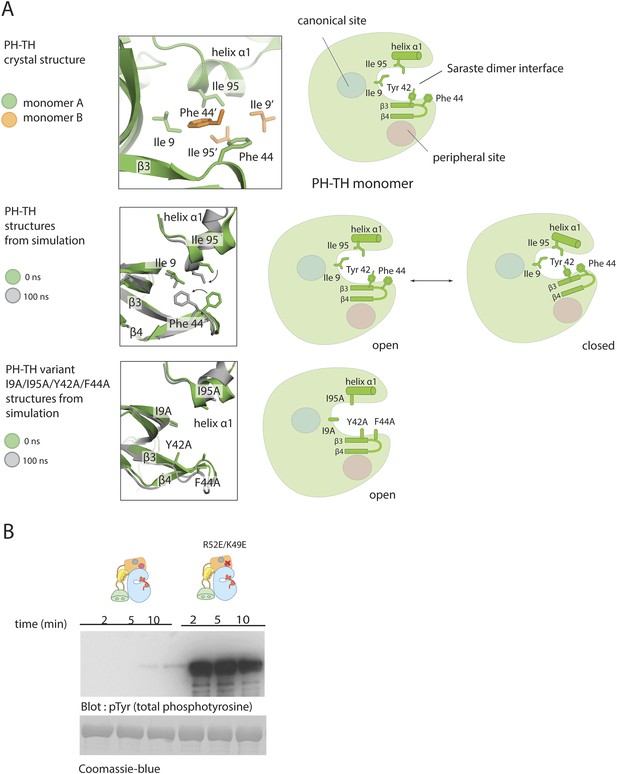
Possible allosteric and electrostatic effects of binding IP6.
(A) Fluctuations of the PH-TH module during a molecular dynamic simulation. An instantaneous structure (t = 100 ns) from a 100 ns simulation of the PH-TH module shows a conformational change that closes the Saraste dimer interface (middle panel). Replacing these hydrophobic residues (IIe 9, Tyr 42, Phe 44 and Ile 95) with alanine prevents the dimer interface from closing in the simulations (lower panel). Although the β3/β4 loop becomes more dynamic, it remains open (see Figure 9–figure supplement 1). Binding of IP6 may shift the equilibrium between open and closed conformations. (B) Activation of the Btk R52E/K49E mutant (2 μM). Arg 52 and Lys 49 are two residues in the peripheral binding site. Mutation of these two residues to glutamate reduces the local charge at the peripheral site by a net four units, and results in Btk activation.
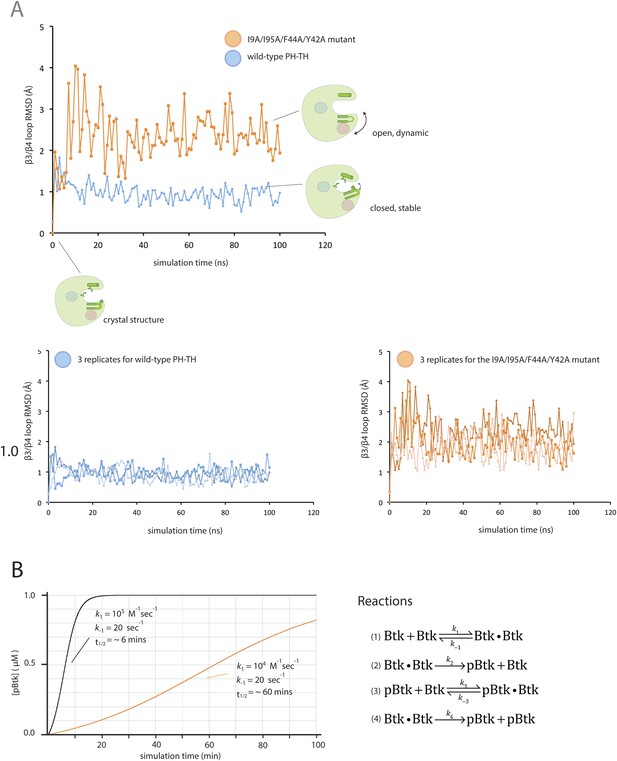
Flexibility in the PH domain of Btk and simulation of the kinetics of Btk autophosphorylation.
(A) Fluctuations in the β3/β4 loop of the wild-type PH-TH module and its I9A/Y42A/F44A/I95A variant. Each trajectory was sampled every 1 ns. The instantaneous structures are aligned using the crystal structure as the reference. The r.m.s. deviation was calculated for the backbone of residues 36 through 57 of the PH-TH domain. (B) Simulations of the kinetics of Btk autophosphorylation. We consider a very simple two-step Michaelis–Menten model for autophosphorylation, in which two Btk molecules first form an encounter complex, followed by phosphorylation of one of the two molecules in the complex. We assume that the rate of complex formation increases if IP6 is bound, and that the catalytic rate constant for phosphorylation of the other increases if one of the two Btk molecules is phosphorylated. This model is described by the following reaction scheme:
The on-rate for a diffusion-limited encounter between two proteins is in the range of 108 to 107 M−1s−1. We assume that in the absence of IP6 the on-rate for Btk dimer formation is much slower than this (k1 = k3 = 104 M−1s−1) and that IP6 binding increases the on-rate by 10-fold (k1 = k3 = 105 M−1s−1). The dissociation rate constant is chosen so that the dissociation constant is 2 mM and 200 μM in the absence and presence of IP6, respective (k-1 = k-3 = 20 s−1). The catalytic constant (k4) for activated Btk is set to 1.0 s−1, comparable to the values determined for other tyrosine kinases, such as Src and ZAP70 (Mukherjee et al., 2013). We assume that the catalytic constant is 10-fold smaller for unphosphorylated Btk (k2 = 0.1 s−1). To compare with our experimental data, the initial concentration of Btk was set at 1 μM. The integration of the differential kinetic equations was done using BerkeleyMadonna (http://www.berkeleymadonna.com/). As shown in the diagram, the autophosphorylation kinetics generated by this simple reaction scheme is consistent with the experimentally observed kinetics.
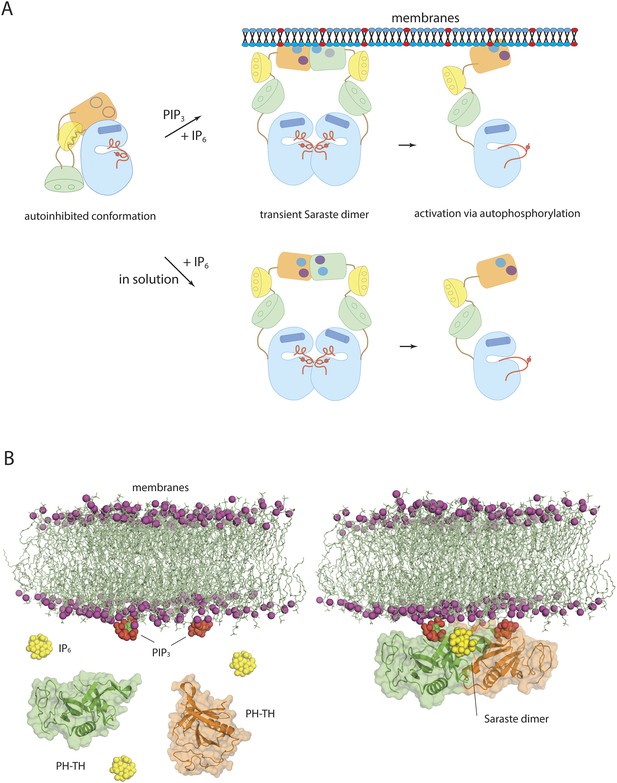
Models of Btk autoinhibition and activation.
(A) The PH-TH module stabilizes the assembled conformation of the Src-like module of Btk in its inactive conformation. The activation of Btk involves formation of a transient dimer through the PH-TH module, which promotes trans-autophosphorylation and activates Btk. The stimulation of dimerization could occur at the membrane or in solution, as in our experiments. (B) IP6 may stimulate membrane-tethered Btk. Formation of the Saraste dimer is compatible with the simultaneous engagement of PIP3 by both PH domains in the dimer, and is compatible with the binding of IP6 at the peripheral binding site. This raises the possibility that IP6 can stimulate dimerization of the membrane-tethered Btk.
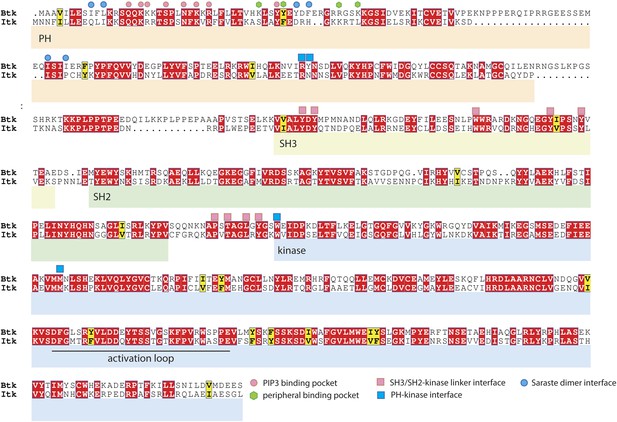
Sequence alignment of Btk and Itk.
The alignment was performed using ClustalW2 (Larkin et al., 2007) and visualized by Espript3 (Robert and Gouet, 2014). The alignment shows that the adoption of the compact autoinhibited conformation by the Src-like module, seen in our crystal structures, is likely to be conserved in Itk. In contrast, residues at the Saraste dimer interface and at the peripheral IP6 binding site in Btk are not conserved in Itk.
Tables
Data collection and refinement statistics
| Src-like module of mouse Btk (217-659) | PH-TH-kinase unit of bovine Btk | Btk PH-TH module bound to IP6 | Btk kinase domain with mutations in the activation loop | |
|---|---|---|---|---|
| PDB ID | 4XI2 | 4Y93 | 4Y94 | 4Y95 |
| Data collection | ||||
| KAu(CN)2 | Native | Native | Native | |
| Wavelength (Å) | 0.9474 | 1.000 | 1.000 | 1.000 |
| Space group | P 31 2 1 | P 2 21 21 | P 1 | P 1 |
| a,b,c (Å) | 132.2, 132.2, 107.6 | 78.6, 38.3, 157.6 | 37.2, 64.0, 80.0 | 50.9, 79.0, 79.2 |
| α,β,γ (°) | 90.0, 90.0, 120.0 | 90.0, 90.0, 90.0 | 82.0, 88.8, 89.8 | 90.7, 89.9, 90.0 |
| Resolution (Å) | 43.2–2.6 | 50–1.7 | 43.9–2.3 | 47.9–1.6 |
| Rsym (%) | 8.7 (>100) | 12.2 (79.7) | 6.0 (53.8) | 5.1 (71.7) |
| I/σ(I) | 10.6 (1.1) | 17.0 (3.5) | 10.8 (1.2) | 13.2 (3.2) |
| Completeness (%) | 91.9 (69.6) | 99.2 (98.5) | 94.9 (71.8) | 96.9 (94.7) |
| Redundancy | 4.8 (3.5) | 7.8 (7.4) | 2.0 (1.7) | 24.4 (3.1) |
| Wilson B factor | 72.8 | 15.7 | 52.3 | 18.1 |
| Refinement | ||||
| Resolution | 43.2–2.6 | 50–1.7 | 46.5–2.3 | 47.9–1.6 |
| Reflections | 57,643(30,933) | 53,357 | 30,516 | 157,562 |
| Rfree reflections | 1525 | 2000 | 2025 | 2009 |
| Rwork/Rfree | 0.237/0.252 | 0.167/0.195 | 0.236/0.254 | 0.155/0.187 |
| No. atoms | ||||
| Protein | 3459 | 6970 | 5053 | 8911 |
| Ligands | 2 | 80 | 216 | 961 |
| Water | 0 | 525 | 39 | 737 |
| Average B factors | ||||
| Protein | 114.4 | 27.7 | 62.1 | 22.5 |
| Solvent | N/A | 33.0 | 66.2 | 30.1 |
| Root mean square deviation from ideality | ||||
| Bonds (Å) | 0.006 | 0.005 | 0.003 | 0.014 |
| Angles (°) | 1.11 | 0.977 | 0.764 | 1.571 |
| Ramachandran statistics | ||||
| Favored (%) | 91 | 98.58 | 98.2 | 97.11 |
| Disallowed (%) | 2.4 | 0.0 | 0.0 | 0.18 |
| MolProbity clash score | 9.6 | 2.8 | 5.16 | 2.66 |
-
The CC1/2 values for the PH-TH-kinase dataset, IP6-bound PH-TH dataset and the kinase domain with mutations in the activation loop dataset are 99.9 (86.5), 99.9 (55.4) and 99.9 (90.7), respectively.
Data statistics for the Src-like module of Btk
| Src-like module of mouse Btk (217-659) | ||||
|---|---|---|---|---|
| Data collection | ||||
| KAu(CN)2 | DMA | Au2(NO3)3 | native | |
| Wavelength (Å) | 0.9474 | 0.9474 | 0.9474 | 0.9474 |
| Space group | P31 2 1 | P31 2 1 | P31 2 1 | P31 2 1 |
| a,b,c (Å) | 132.2, 132.2, 107.6 | 132.5, 132.5, 107.3 | 131.9, 131.9, 107.6 | 131.8, 131.8, 107.0 |
| α,β,γ (°) | 90.0, 90.0, 120.0 | 90.0, 90.0, 120.0 | 90.0, 90.0, 120.0 | 90.0, 90.0, 120.0 |
| Resolution (Å) | 43.2–2.6 | 41.7–3.5 | 41.7–3.4 | 40–4.0 |
| Rsym (%) | 8.7(>100) | 8.7 (32.7) | 7.5 (32.5) | 5.1 (71.7) |
| I/σ(I) | 10.6 (1.1) | 9.7 (3.0) | 12.1 (3.6) | 18.4 (12.3) |
| Completeness (%) | 91.9 (69.6) | 92.4 (94.2) | 94.5 (95.9) | 92.3 (100) |
| Redundancy | 4.8 (3.5) | 4.3 | 4.1 | N/A |
| Wilson B factor | 72.8 | 58.8 | 70.8 | 67.3 |
Thermodynamic parameters for Btk PH-TH module binding to IP6 and IP4
| Protein | Ligand | N | Ka (×106) | Kd (nM) | ΔH (Kcal/mol) | ΔS (cal/mol/deg) |
|---|---|---|---|---|---|---|
| Wild-type PH-TH | IP6 | 1.1 ± 0.1 | 4.2 ± 0.3 | 238 ± 32 | −1.1 ± 0.2 | 26.7 |
| Wild-type PHTH | IP4 | 0.9 ± 0.1 | 39 ± 11 | 26 ± 6 | 4.4 ± 0.1 | 49.5 |
| PH-TH R28C/D24N | IP6 | 0.7 ± 0.2 | 0.21 ± 0.03 | 4760 ± 700 | −1.7 ± 0.6 | 17.5 |
-
The integrated heat from representative isothermal titration calorimetry experiments was fit with a one-binding site model. See methods for the details of data analysis.





