Kinetochore-independent chromosome segregation driven by lateral microtubule bundles
Figures
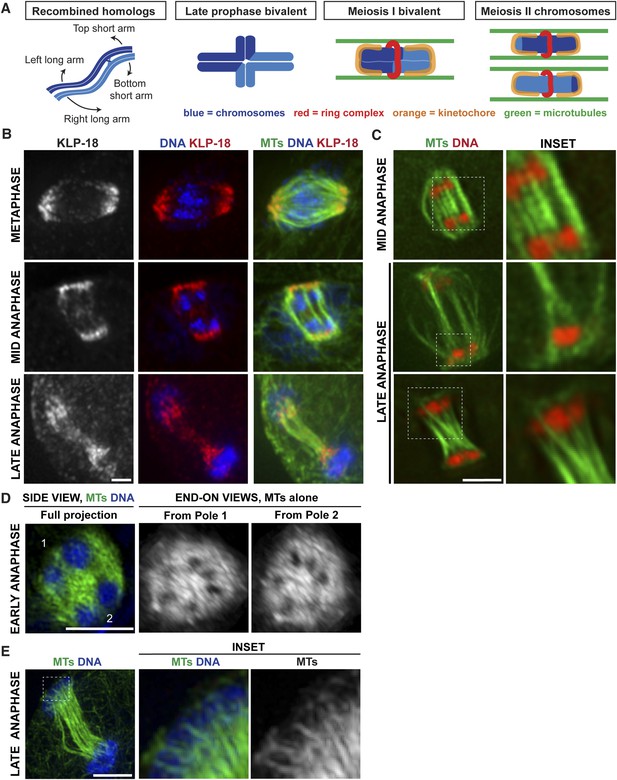
Poles broaden but remain intact during anaphase, creating open channels.
(A) Chromosome organization during C. elegans meiosis. Homologous chromosomes (blue) undergo recombination to link them together and then reorganize to form a cruciform bivalent with long and short arms. This structure condenses further and a holocentric cup-like kinetochore (orange) forms around the bivalent ends and a complex of proteins (red) forms a ring around the center. Following homolog separation, the two resulting chromosomes each form a similar structure, with kinetochores cupping the chromosome ends and a ring around the sister chromatid interface. Bundles of microtubules (green) associate laterally with chromosomes during both meiotic divisions. (B) Oocyte spindles stained for tubulin (green), DNA (blue), and spindle pole marker KLP-18 (red). KLP-18 is at focused poles in metaphase (top) and remains as poles broaden in mid-anaphase (middle). Chromosomes move past the poles in late anaphase (bottom). (C) Live imaging of worms expressing mCherry-histone (red) and GFP-tubulin (green). Full projections of entire spindles are shown on the left and partial projections of insets chosen to highlight particular features of spindle organization are on the right. Lateral bundles associate with and extend beyond segregating chromosomes in mid-anaphase (top row) and maintain association with chromosomes as they move past the poles (bottom). (D) Super-resolution image of an early anaphase oocyte spindle stained for DNA (blue) and tubulin (green). End-on views show a rotated 3D rendering and highlight the six open channels between segregating chromosomes. (E) Super-resolution image of a late anaphase oocyte spindle stained for DNA (blue) and tubulin (green) showing microtubule bundles splitting around the chromosomes as they reach microtubule ends. Bars = 2.5 μm.
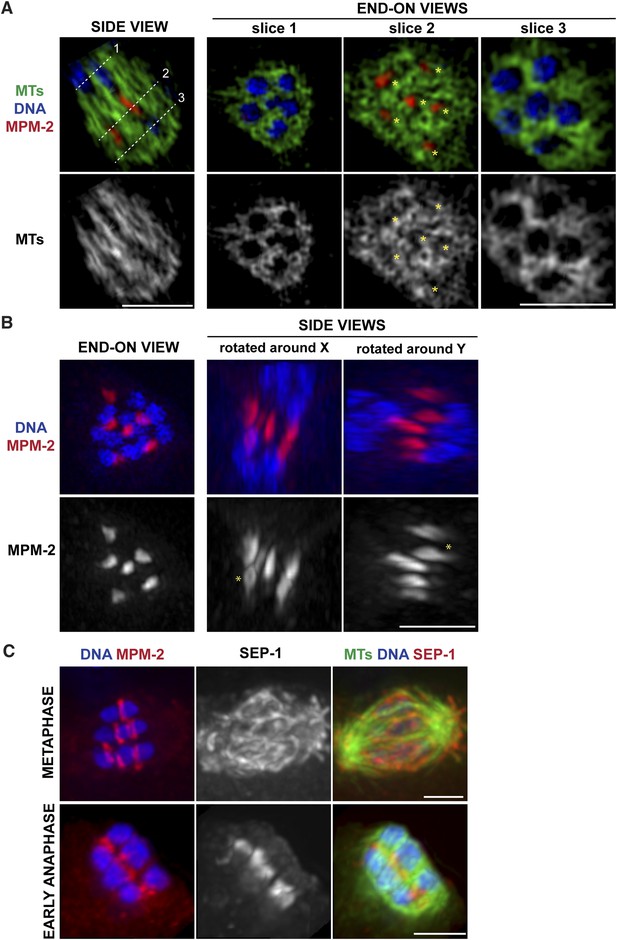
Ring structures are located within the microtubule channels during anaphase.
(A, B) Super-resolution imaging of an anaphase oocyte spindle stained for DNA (blue), tubulin (green), and ring marker MPM-2 (red). (A) The first column shows a single z-slice highlighting an open channel containing a separating chromosome pair with a flattened ring in between. Columns 2, 3, and 4 are end-on views (each a single z-slice, depicting the region denoted by the corresponding dotted line on the first image), showing the chromosomes and disassembling rings within microtubule channels. (B) Rings dissociate from the chromosomes; although most of the structures appear flattened, partial rings can still be observed (‘asterisks’). (C) Oocyte spindles stained for DNA (blue), ring marker MPM-2 (red in left column), SEP-1/separase (middle column, red in right column), and tubulin (green). SEP-1 localizes to cup-like structures and filaments within the spindle during metaphase, consistent with kinetochore localization (top row). In anaphase (bottom row), SEP-1 localizes to the rings. Bars = 2.5 μm.
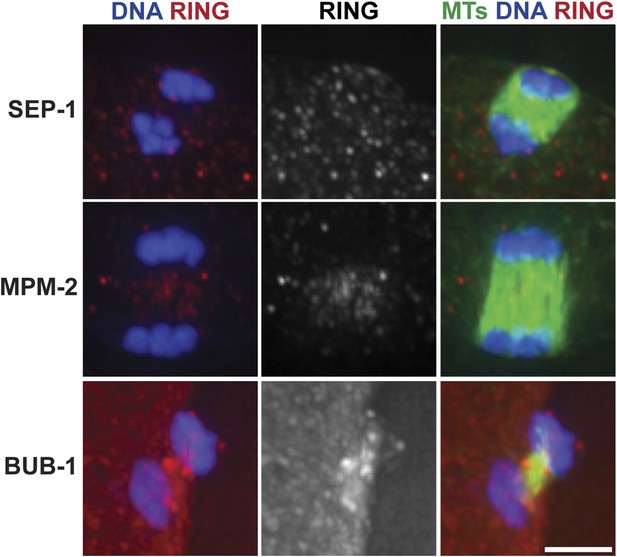
Ring structures are gone in late anaphase.
Late anaphase oocyte spindles stained for DNA (blue), ring markers (SEP-1, MPM-2, BUB-1, respectively; red), and tubulin (green). At this stage, ring components are no longer detectable in distinct structures. Bar = 2.5 μm.
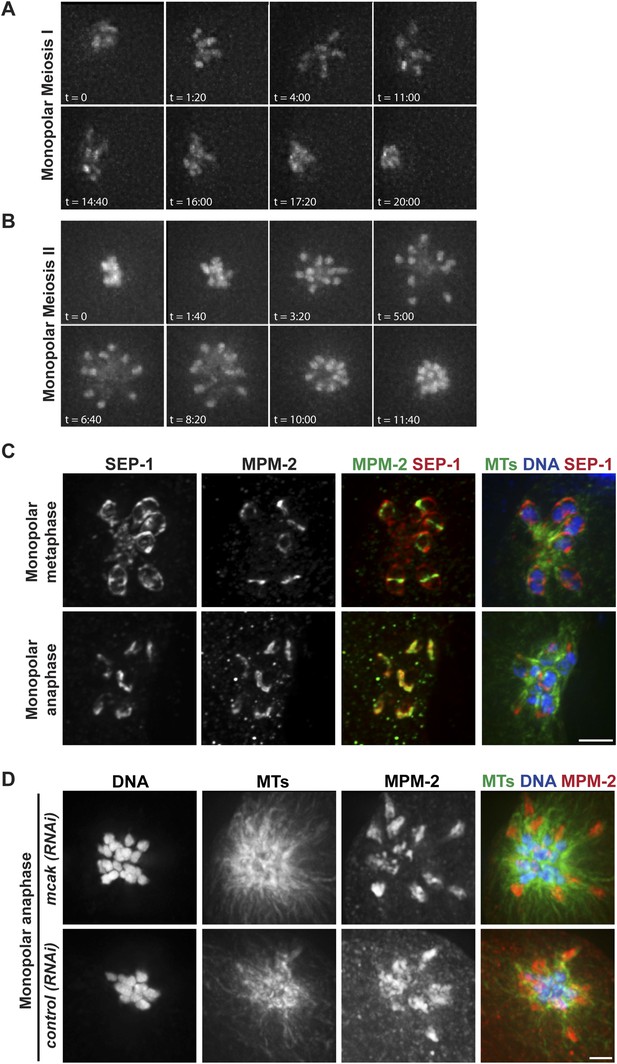
Chromosomes move towards microtubule minus ends during monopolar anaphase.
(A, B) Montages of time-lapse imaging of a strain expressing GFP-histone; GFP-tubulin following klp-18 (RNAi) to generate monopolar spindles during Meiosis I (A) and II (B). Chromosomes move away from the pole during monopolar spindle formation and then coordinately move polewards during anaphase. (C) klp-18 (RNAi) monopolar oocyte spindles stained for DNA (blue), SEP-1 (red), ring marker MPM-2 (green in third column), and tubulin (green in fourth column). When chromosomes are away from the pole in metaphase, SEP-1 localizes to kinetochores and then moves to the ring structures in anaphase. (D) klp-18 (RNAi) monopolar oocyte spindles stained for tubulin (green), DNA (blue), and ring marker MPM-2 (red). In the top row, MCAK/KLP-7 was also depleted, creating a larger spindle. When separated chromosomes move to the pole in monopolar anaphase, the rings stay out. Bars = 2.5 μm.
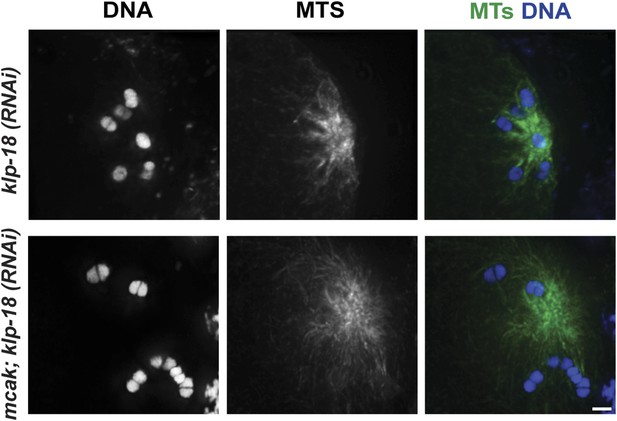
MCAK inhibition causes larger monopolar spindles.
Monopolar oocyte spindles generated by RNAi-mediated depletion of KLP-18, stained for DNA (blue) and microtubules (green). The top panel shows a normal klp-18(RNAi) monopolar spindle, and the bottom panel highlights the increase in spindle size following RNAi depletion of MCAK/KLP-7. Bar = 2.5 μm.
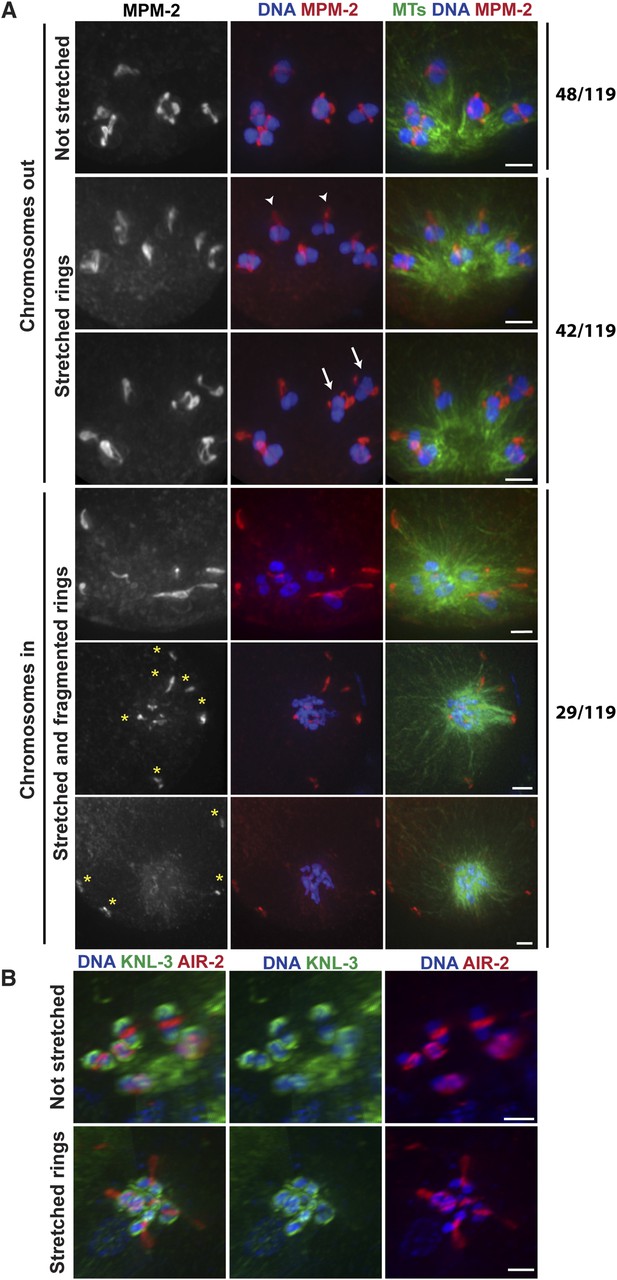
Chromosomes are subjected to minus-end directed forces prior to anaphase.
(A) Monopolar oocyte spindles arrested at Metaphase I stained for DNA (blue), tubulin (green), and ring marker MPM-2 (red). Top row shows intact rings. Rows 2 and 3 show examples of rings stretching, but still near chromosomes (‘arrowheads’ show rings predominantly coming off one side of the bivalent, ‘arrows’ show rings off both sides). Bottom three rows show examples of chromosomes at the pole and ring fragments (‘asterisks’) at microtubule plus ends or off the spindle. (B) Monopolar oocyte spindles arrested at Metaphase I stained for DNA (blue), kinetochore protein KNL-3 (green), and ring component AIR-2 (red). The kinetochore remains intact in spindles with stretched rings, indicating that the cells have not initiated anaphase. Bars = 2 μm.
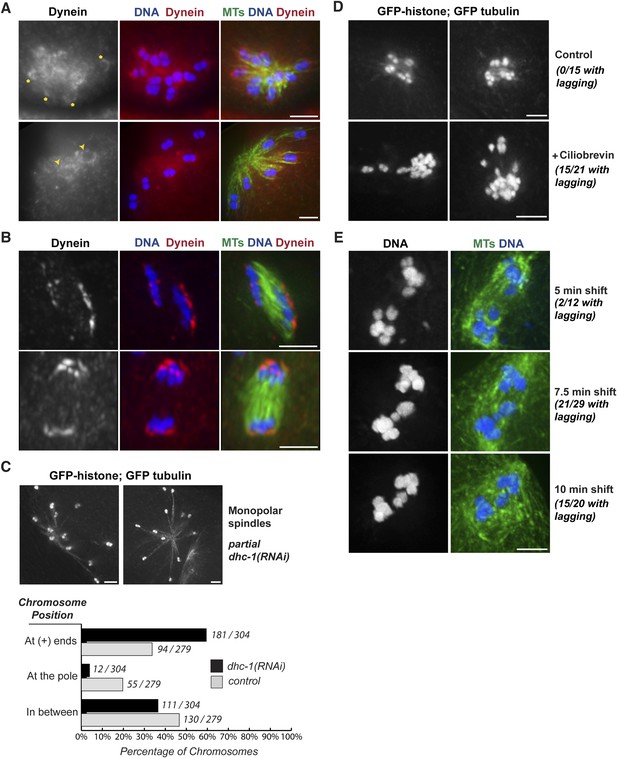
Dynein contributes to chromosome segregation on oocyte spindles.
(A) Monopolar oocyte spindles stained for tubulin (green), DNA (blue), and DHC-1/dynein (red). The tubulin and DNA channels were deconvolved, but the dynein channel was not, to show a faint pool of dynein surrounding chromosomes. Dynein cups chromosomes, sometimes found enriched on the side facing away from the pole (‘asterisks’) and sometimes around the entire chromosome (‘arrowheads’); partial projections are shown to highlight particular chromosomes. (B) Dynein localization on anaphase spindles with the same colors as (A), except the dynein channel was deconvolved. Dynein localizes adjacent to the outside surface of separating chromosomes. (C) Images of monopolar spindles in a strain expressing GFP-histone; GFP-tubulin following partial dynein RNAi. Spindles are larger with chromosomes often located at the extreme ends microtubules; graph depicts chromosome positions on monopolar spindles in dhc-1 (RNAi) (black bars) compared to control (gray bars). (D) Images of monopolar anaphase spindles in a strain expressing GFP-histone; GFP-tubulin with no treatment (top row) or following incubation with 5 μM ciliobrevin for 20 min (bottom row). Following dynein inhibition, lagging chromosomes were observed in a significant fraction of spindles (15/21). (E) Images of bipolar anaphase spindles following dynein inhibition; DNA is shown in blue and microtubules in green. The dynein temperature-sensitive mutant dhc-1(ct76ts) was shifted to the restrictive temperature for 5 (top), 7.5 (middle), or 10 min (bottom); spindles with lagging chromosomes were observed under all three conditions. Bars = 2.5 μm.
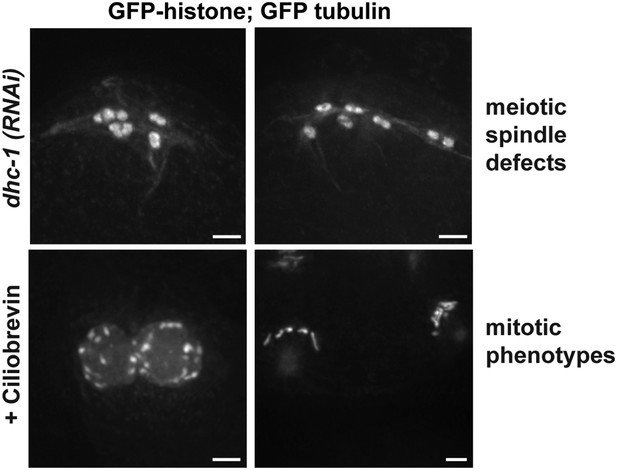
Strong dynein inhibition causes spindle defects.
Images of a strain expressing GFP-histone and GFP-tubulin to show chromosomes and microtubules, respectively. Top row: Strong dhc-1 (RNAi) causes defects in oocyte spindle morphology. Bottom row: Incubation of worms in high concentrations of the dynein inhibitor ciliobrevin (30μM for 30 minutes) leads to mitotic defects consistent with dynein inhibition, such as errors in pronuclear fusion (bottom left; [Gonczy et al., 1999]), and monopolar mitotic spindles (bottom right; [Vaisberg et al., 1993]), showing that ciliobrevin works to inhibit dynein in C. elegans. Bars = 2.5 μm.
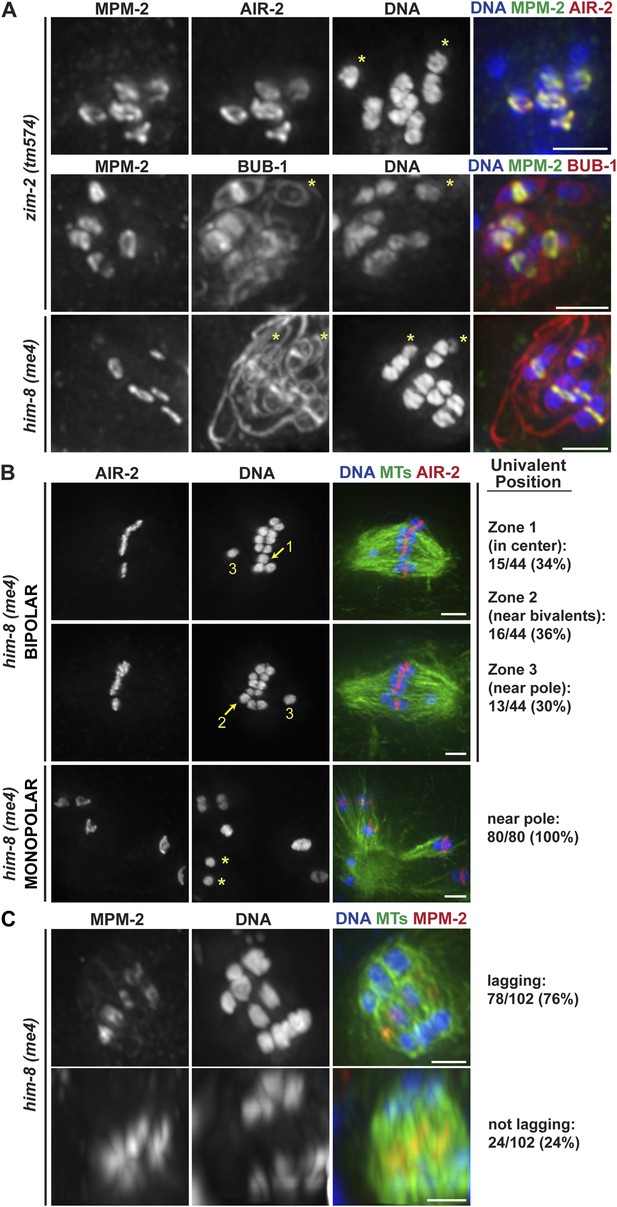
Univalents lacking rings exhibit congression and segregation errors.
(A) Oocytes from him-8(me4) and zim-2(tm574) worms stained for DNA (blue), AIR-2 or BUB-1 (red), and MPM-2 (green); each of these strains has two univalents (‘asterisks’). Rings are not detected on univalents, but kinetochore staining is present (BUB-1; bottom two rows). (B) Metaphase-arrested him-8(me4) bipolar and monopolar spindles, stained for DNA (blue), AIR-2 (red), and tubulin (green). During congression, univalents are randomly positioned on the bipolar spindle (top two rows), but were located near the pole on every monopolar spindle we observed (bottom). For quantification of bipolar spindles, univalents were scored as being at the center of the metaphase plate (zone 1), partially overlapping with the bivalents (zone 2), or not overlapping (zone 3). Examples of each category are indicated on the images. (C) him-8(me4) anaphase spindles stained for DNA (blue), MPM-2 (red), and tubulin (green). During segregation, univalents are either found near other segregating chromosomes (bottom) or lag behind in the center (top). Bars = 2.5 μm.
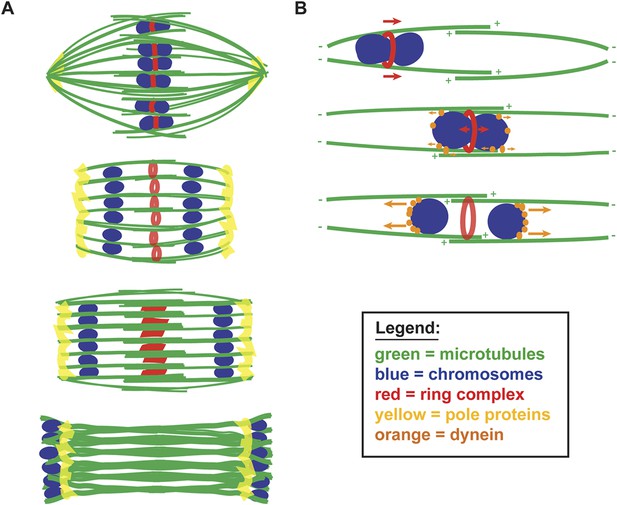
A balance of forces controls chromosome movement on the acentrosomal spindle.
(A) Model depicting microtubules (green), chromosomes (blue), rings (red), and spindle pole proteins (yellow). During metaphase, poles are focused and chromosomes are aligned between overlapping microtubule bundles. During anaphase, poles broaden creating open channels and separated chromosomes move along lateral bundles until they are past the poles; at this stage, the channels close. In anaphase, rings are removed from chromosomes and then flatten and disassemble. (B) Model depicting microtubules (green), chromosomes (blue), rings (red), and dynein (orange). During congression, KLP-19 and potentially other ring components provide a plus-end directed force (‘red arrows’), moving the chromosome toward the metaphase plate; during this time dynein begins to accumulate on chromosomes, resulting in weak minus-end forces (‘small orange arrows’). At metaphase, the plus- and minus-end directed forces are each balanced as the chromosome associates with microtubules in an overlap zone. Ring removal at the metaphase to anaphase transition shifts this balance, triggering poleward movement. In anaphase, more dynein has accumulated on chromosomes, resulting in stronger minus-end forces (‘large orange arrows’).
Videos
Super-resolution imaging revealing six open channels in the early anaphase spindle.
Super-resolution image of a fixed oocyte spindle stained for tubulin. Video is a rotation of a 3D volume rendering generated using Softworx software (Applied Precision).
Super-resolution view of spindle organization in mid-anaphase.
Super-resolution image of a fixed oocyte spindle stained for DNA (blue), tubulin (green), and ring marker MPM-2 (red). Video shows a 3D volume rendering, generated using Imaris software (Bitplane) and the Orthogonal Slice View function is used to show individual slice views within the spindle. Individual components blink in and out to highlight particular features of spindle organization; specifically that each pair of separating chromosomes is located within an open microtubule channel, with a disassembling ring structure in between.
Super-resolution view of spindle organization in mid-anaphase.
Super-resolution image of a fixed oocyte spindle stained for DNA (blue), tubulin (green), and ring marker MPM-2 (red). This is the same video as Video 2, but without individual components blinking in and out.
Super-resolution view of spindle organization in mid-anaphase.
Super-resolution image of a fixed oocyte spindle stained for DNA (blue) and tubulin (green). This is the same sequence as Video 3, but with only DNA and tubulin displayed.
Super-resolution view of spindle organization in mid-anaphase.
Super-resolution image of a fixed oocyte spindle stained for tubulin (green) and ring marker MPM-2 (red). This is the same sequence as Video 3, but with only tubulin and the rings displayed.
Dynamics of monopolar Meiosis I.
Time-lapse imaging of a strain expressing GFP-histone; GFP-tubulin following klp-18 (RNAi) to generate monopolar spindles. In monopolar Meiosis I, the 6 bivalents move away from the central pole, oscillate, and then coordinately move inwards to the pole. Frames were acquired every 10 s and displayed at 5 frames per second.
Dynamics of monopolar Meiosis II.
Time-lapse imaging of a strain expressing GFP-histone; GFP-tubulin following klp-18 (RNAi) to generate monopolar spindles. In monopolar Meiosis II, the 12 separated chromosomes move away from the central pole, oscillate, and then coordinately move inwards to the pole. Frame acquisition same as Video 6.
Additional files
-
Supplementary file 1
Complete list of strains used in this study.
- https://doi.org/10.7554/eLife.06462.020





