MicroRNA-203 represses selection and expansion of oncogenic Hras transformed tumor initiating cells
Figures
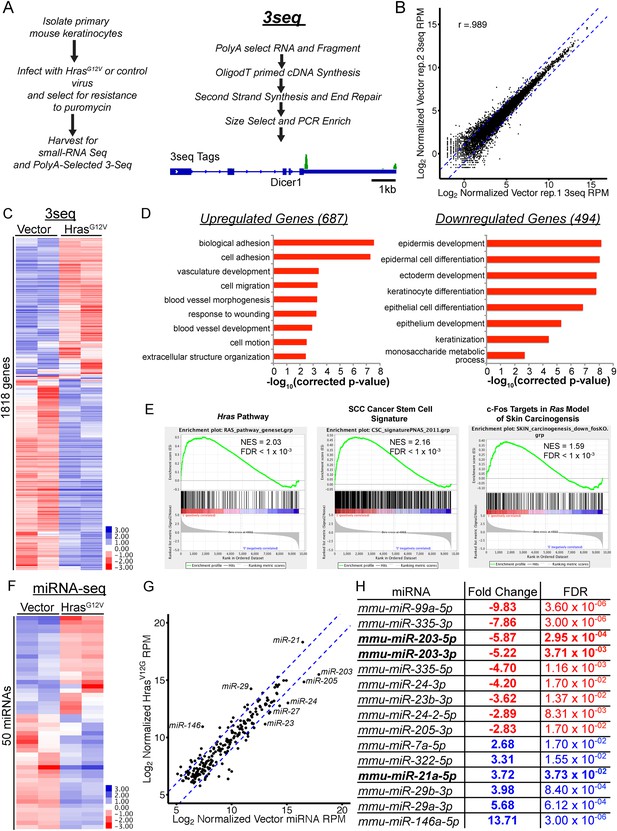
Genome-wide profiling of the oncogenic HrasG12V-transformed miRNA and mRNA transcriptome in primary keratinocytes.
(A) Schematic of experimental approach to identify deregulated mRNA and miRNA networks driven by oncogenic HrasG12V using small-RNA Seq and 3Seq. The 3seq library preparation allows quantitative definition of poly-A+ RNA 3′ends and expression levels. (B) 3Seq reproducibly detects mRNA expression levels over 4 orders of magnitude. Pearson correlation coefficient displayed (C) unsupervised hierarchical clustering of log-transformed mean-centered mRNA expression levels for all transcripts deregulated twofold by oncogenic HrasG12V (n = 2 libraries per condition) (D) Gene Ontology analysis of transcripts up and downregulated by HrasG12V (twofold change FDR <0.05) indicates enrichment for migratory and angiogenic processes, and suppression of keratinocyte differentiation. (E) GSEA analysis of selected genesets relevant to skin carcinogenesis. (F) Unsupervised hierarchical clustering of log-transformed mean-centered miRNA expression levels for all transcripts deregulated twofold by oncogenic HrasG12V (n = 2 libraries per condition) (G, H) Abundant miRNAs such as miR-203, miR-205, and miR-21 are strongly deregulated by oncogenic Ras.
-
Figure 1—source data 1
Log2 fold changes for transcripts up or down regulated twofold with FDR <0.05 in HrasG12V-transformed keratinocytes.
- https://doi.org/10.7554/eLife.07004.004
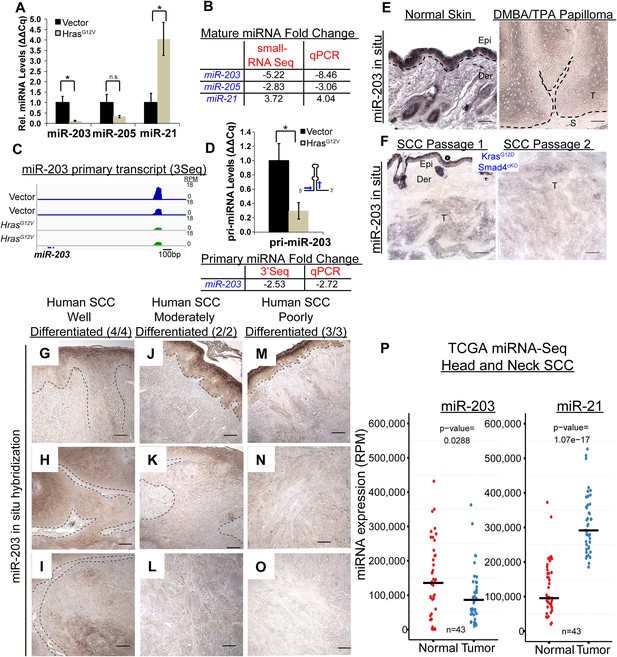
miR-203 is strongly suppressed in mouse and human SCCs.
(A, B) qPCR and small-RNA-Seq independently validate downregulation of miR-203 and upregulation of miR-21 driven by oncogenic HrasG12V (n = 3 biological rep. qPCR, n = 2 small-RNA-Seq, mean ± SEM displayed, *p < 0.05, Student's t-test two-sided). (C) Gene track and quantification the 3′end of the miR-203 primary transcript based on 3Seq. (D) miR-203 primary transcript detection by qPCR (n = 3 biological replicates, Mean ± SEM displayed, *p < 0.05, ns = non-significant, Student's t-test two-sided). (E) miR-203 is downregulated in DMBA/TPA produced papillomas compared to normal adjacent tissue. Epi = epidermis, Der = dermis, T = tumor, and S = stroma. The black lines denote the epidermal/dermal and tumor/stroma boundary (F) miR-203 is downregulated in malignant SCCs derived from KrasG12D/Smad4cKO and passaged in immunocompromised mice. (G–O) Reduced miR-203 expression is correlated with increasing malignancy in human skin SCC cancers. Panels G, J, M were taken from regions with more histologically normal regions to demonstrate successful miR-203 hybridization. (P) miRNA-Seq quantification from patient matched normal and tumor tissue obtained from the TCGA consortium data (bar indicates mean value, Student's t-test two-sided). Scale bar = 50 μm.
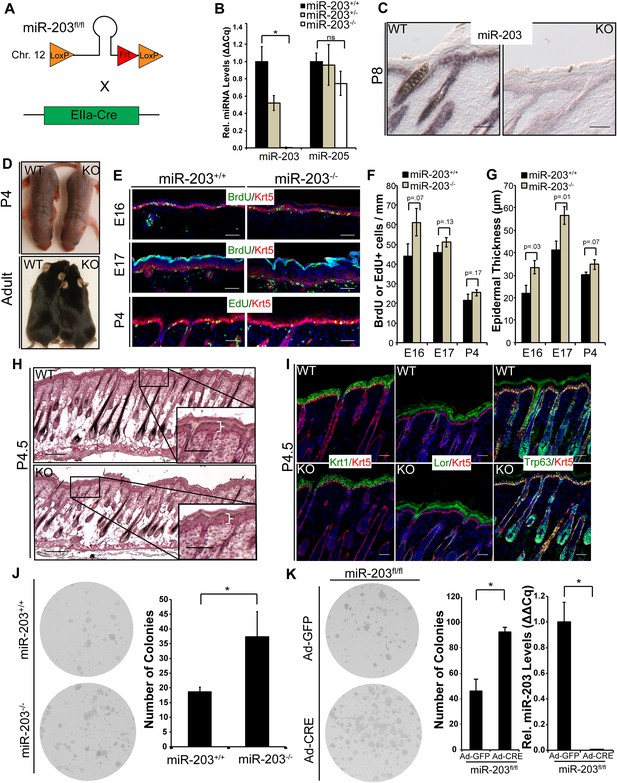
Loss of miR-203 modestly impairs embryonic epidermal development.
(A) Schematic of miR-203 conditional allele generation and knockout strategy. (B) Validation of miR-203 ablation by qPCR from isolated epidermal samples (n = 3 biological replicates, * p < 0.05, ns = non-significant, Student's t-test two-sided). (C) Validation of miR-203 ablation within the epidermis by in situ hybridization (Scale bar = 50 μm). (D) miR-203 knockout mice are visibly indistinguishable from wild-type counterparts. (E–G) miR-203 ablation results in mild epidermal hyperplasia during embryonic development. (n = 3 E16, n = 4 E17, and n = 3 p4 animals, p-value provided in figure, Student's t-test one-sided). (H) Representative hematotoxylin and eosin image from p4.5 animals, demonstrating restored normal skin morphology in neonatal animals. (Scale bars = 50 μm for inset and 100 μm for main images) (I) Epidermal differentiation is not compromised by loss of miR-203. (Scale bars = 50 μm) (J) miR-203−/− primary keratinocytes are more clonogenic than wild-type counterparts (representative results from 3 experiments, *p < 0.05, Student's t-test two-sided). (K) Conditional ablation of miR-203 from passaged miR-203fl/fl keratinocytes results in higher clonogenicity (representative results from n = 3 independent experiments, mean ± standard deviation displayed, *p < 0.05, Student's t-test, two-sided).
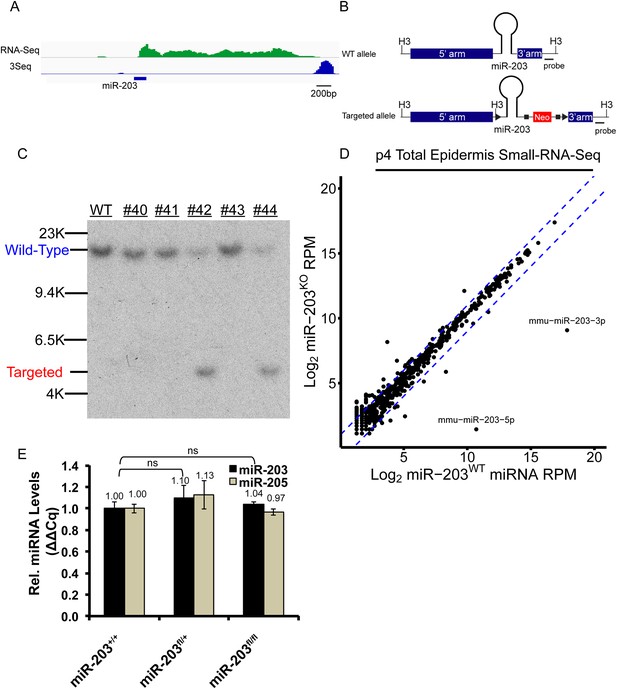
Generation of a miR-203 conditional knockout mouse.
(A) RNA-Seq and 3Seq detection of the miR-203 primary transcript (B) Schematic of the miR-203 conditional allele. H3 = HindIII (C) Southern blot confirmation of founder miR-203 conditional knockout mice (D) Small-RNA-seq confirmation of miR-203 loss. Blue lines indicate twofold change (E) qPCR quantification of miR-203 and miR-205, demonstrating that the miR-203floxed allele does not alter microRNA levels. Error bars represent S.E.M from reactions performed from n = 2 mice in technical triplicate, ns = non-significant.
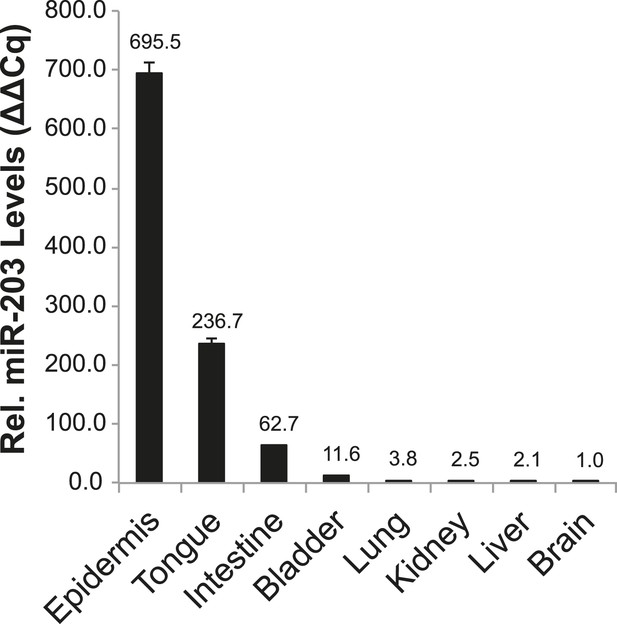
miR-203 expression in diverse mouse tissues.
qPCR detection of mature miR-203 in various adult mouse organ tissues. Error bars represent S.E.M from reactions performed in technical duplicate.
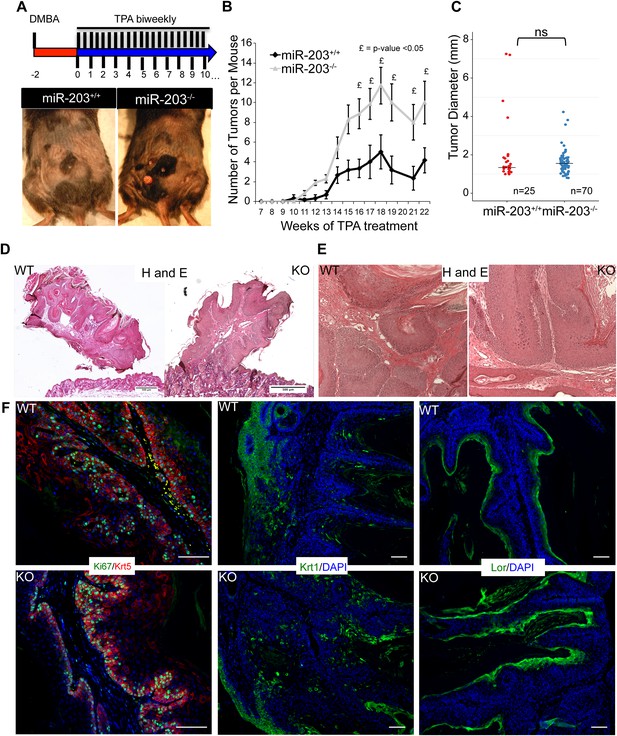
Loss of miR-203 sensitizes mice to DMBA/TPA skin carcinogenesis.
(A) Representative images of tumors that were formed in the skin of WT and miR-203 null mice treated with DMBA/TPA. (B) miR-203−/− mice have a larger tumor burden than miR-203+/+ counterparts (n = 6 and 7 miR-203+/+ and miR-203−/− animals respectively, mean ± SEM displayed, £ = p < 0.05, Whitney–Mann U-test one-sided). (C) miR-203−/− tumor size distribution is similar to wild-type animals (ns = non-significant, Student's t-test two sided, median displayed as bar). (D, E) miR-203+/+ and miR-203−/− papillomas display similar morphologies and histology. (F) Proliferation and differentiation dynamics are similar between miR-203+/+ and miR-203−/− tumors. (Scale bars = 50 μm).
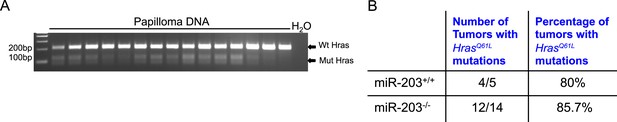
The HrasQ61L mutation is common in both miR-203+/+ and miR-203−/− tumors.
(A) Representative end-point PCR followed by XbaI digestion. The HrasQ61L allele produces 120 bp and 87 bp product, whereas wild-type produces a 207-bp product (B) Table with quantification of genotyping results. non-significant p > 0.05, chi-squared test.
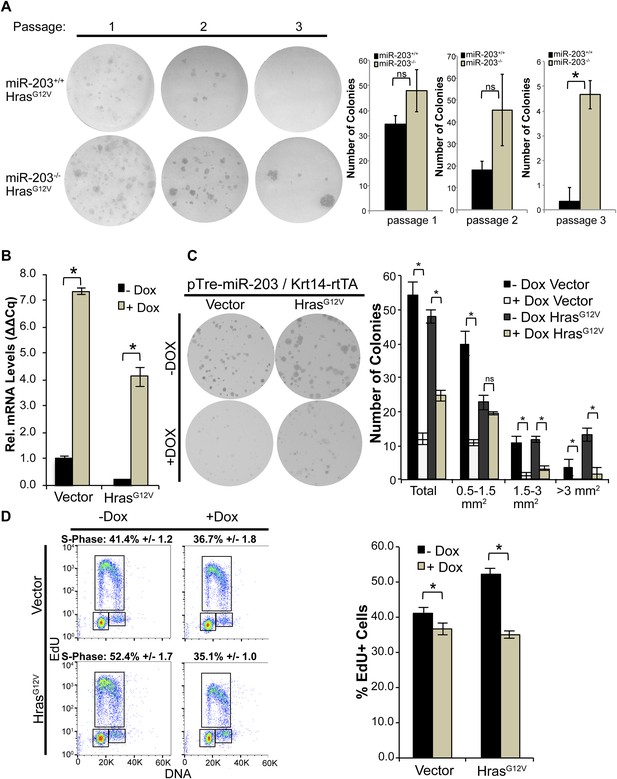
miR-203 antagonizes HrasG12V-driven keratinocyte proliferation.
(A) HrasG12V transduced miR-203−/− primary cultures are more colonogenic upon serial passage than wild-type controls (representative of n = 2 independent experiments, mean ± standard deviation displayed. *p < 0.05, ns = non-significant, Student's t-test two-sided). (B) qPCR of miR-203 induction upon addition of doxycycline in vector and HrasG12V transduced cells (mean ± SEM displayed, n = 3 biological replicates). (C) Restoration of miR-203 using a doxycycline-inducible transgene results in suppression of colony formation ability in HrasG12V transduced and control keratinocytes. miR-203 was induced with doxycycline (5 μg/ml) 24 hr after plating (representative of n = 3 independent experiments, mean ± standard deviation displayed, *p ≤ 0.05, , ns = non-significant). (D) miR-203 restoration suppresses HrasG12V-driven S-Phase entry. miR-203 was induced for 24 hr prior to harvesting for flow cytometry. (n = 3, mean ± standard deviation displayed, *p ≤ 0.05).
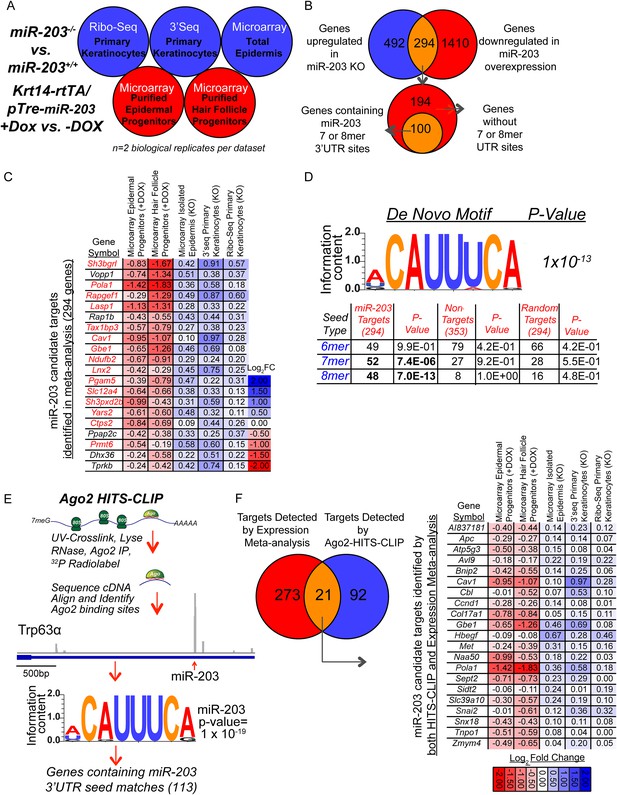
Comprehensive identification of miR-203 targets using genome-wide expression analyses and Ago2 HITS-CLIP.
(A) Schematic of genome-wide expression profiling data sets used in meta-analysis to identify bono-fide miR-203 targets. (B) Genes upregulated in all three miR-203 loss-of-function data sets (786 genes, no fold-change or p-value cut-off) were compared to genes downregulated in both miR-203 gain-of-function data sets (1704 genes, no fold-change or p-value cut-off) to identify a subset of genes with a strong inverse correlation to miR-203 expression (294 genes) of which 100 genes contained miR-203 7mer or 8mer seed sequence matches in their 3′UTRs. (C) Table demonstrating top 20 genes identified in meta-analysis ranked by negative-correlation to miR-203 expression. Genes colored in red contain 3′ UTR miR-203 7 or 8mer seed matches. (D) De novo motif searching identified an 8mer miR-203 seed motif, complementary to the miR-203 seed sequence enriched in the 3′UTR of candidate miR-203 target genes identified in the meta-analysis (294 genes). Table demonstrating enrichment for 7 or 8mer seed matches in the 3′UTR of candidate miR-203 target genes (294) over the background seed distribution in primary keratinocytes, which is not seen for randomly selected 294 genes expressed in primary keratinocytes or a negative control gene set of genes upregulated in miR-203 gain-of-function and downregulated miR-203 loss-of-function (353 genes). (E) Schematic of Ago2 HITS-CLIP and the identified miR-203 seed motif. (F) Diagram of genes detected by expression meta-analysis and Ago2-HITS-CLIP. Table of 21 high confidence miR-203 targets identified through expression meta-analysis and that have Ago2-HITS-CLIP 3′UTR peaks with miR-203 seed matches.
-
Figure 6—source data 1
GO-analysis of selected miR-203 data sets.
- https://doi.org/10.7554/eLife.07004.013
-
Figure 6—source data 2
Putative miR-203 targets detected in the expression meta-analysis.
- https://doi.org/10.7554/eLife.07004.014
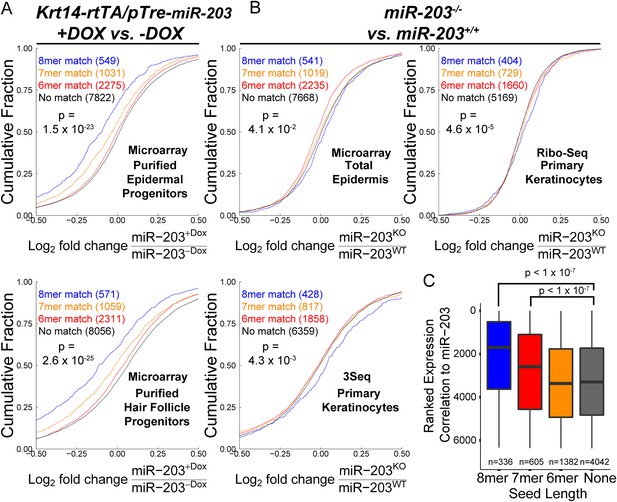
Transcripts containing 3′UTR miR-203 seed matches are regulated by miR-203.
(A) Genes containing miR-203 seed matches are more likely to be downregulated upon miR-203 overexpression or (B) upregulated upon miR-203 ablation (panel B) (p-value <0.05 for comparison of 8mer match to no-match, K–S test, one-sided). (C) Transcripts are ranked based upon aggregate fold-changes consistent with miR-203 regulation to produce a ranked expression correlation metric. Transcripts with 7 or 8mer miR-203 seed matches are more likely to be regulated by miR-203 modulation. (p < 0.05) (see ‘Materials and methods’).
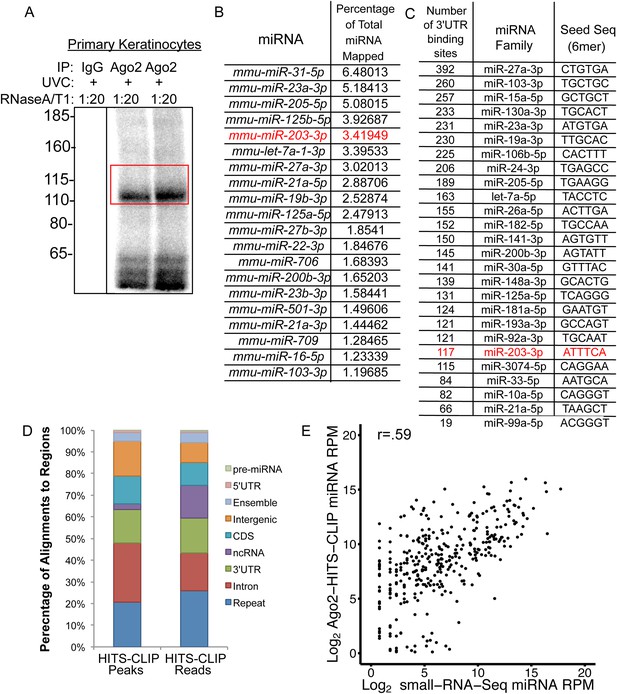
Ago2-HITS-CLIP in primary keratinocytes.
(A) Example autoradiogram of isolated Ago2-RNA complexes, red box indicates region excised for sequencing. (B) Proportion of miRNAs detected by Ago2-HITS-CLIP. (C) Number of 3′UTR Ago2 peak containing seed sequences from miRNA families accounting for 90% of all miRNAs in p4 epidermis. (D) Genome-wide distribution of Ago2 peaks and reads. (E) Comparison of miRNAs detected by Ago2-HITS-CLIP and small-RNA-Seq.
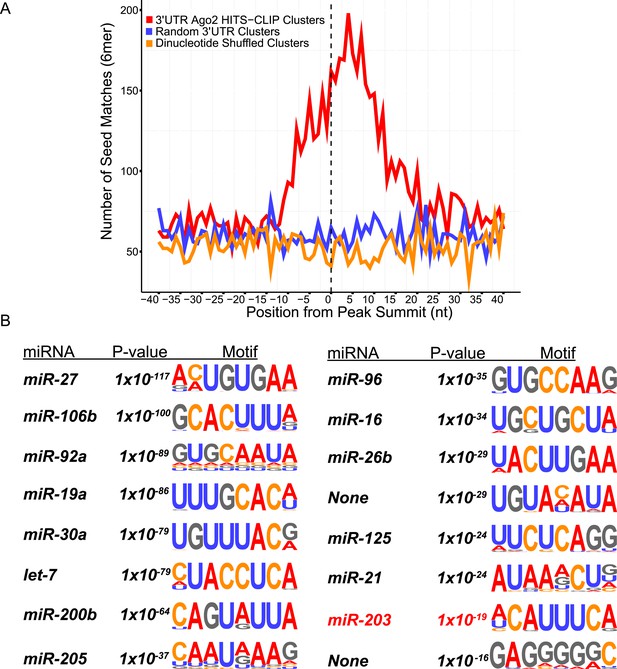
Ago2 HITS-CLIP 3′UTR peaks are enriched in keratinocyte miRNA seed matches, including miR-203.
(A) Position of miRNA seed matches for miRNAs highly expressed in total epidermal samples. The peak summit represents nucleotide position 0 (B) De novo motif searching identifies the most enriched 8mer motifs in 3′UTR peaks.
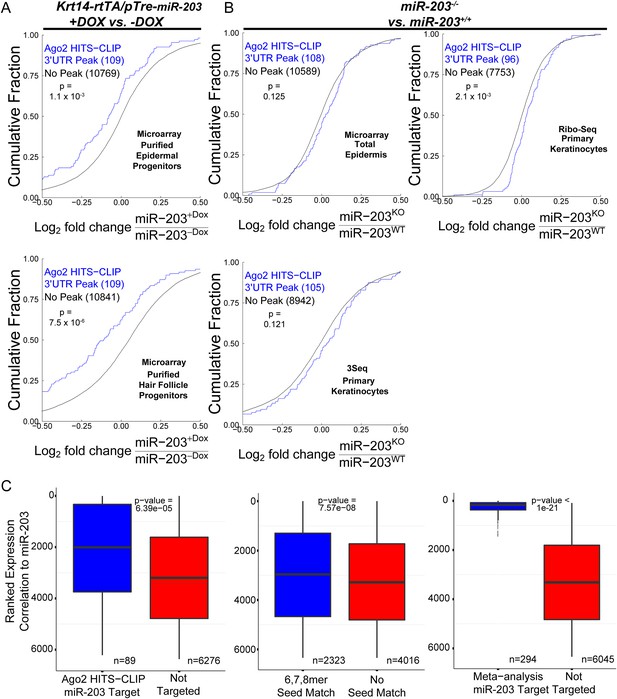
Predicted miR-203 targets based on HITS-CLIP are regulated by miR-203.
(A) Cumulative distributions of miR-203 targets or not targeted transcripts based on HITS-CLIP in miR-203 overexpression data sets. (B) Cumulative distributions of miR-203 targets or not targeted transcripts based on HITS-CLIP in miR-203 knockout data sets (C) Ranked analysis of miR-203 targets detected by HITS-CLIP, based on 6, 7, 8mer seed only, or through meta-analysis (p value displayed on plots).
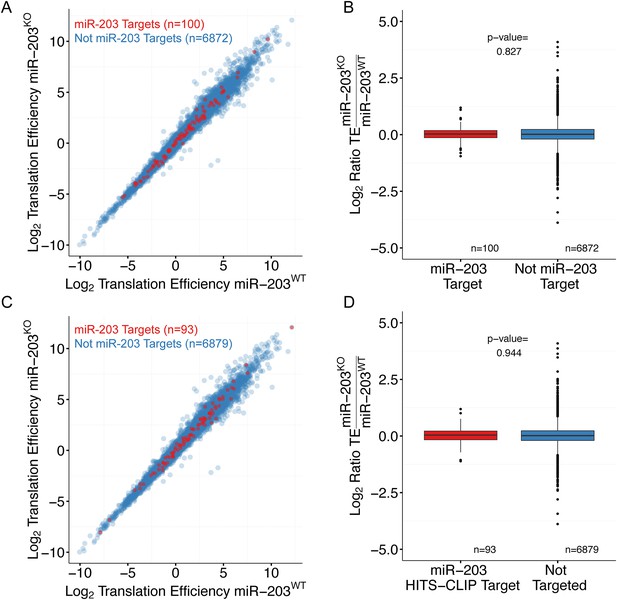
miR-203 targets do not display translation efficiency changes upon miR-203 ablation.
(A) Comparison of translation efficiency for miR-203 targets (red) and non-targeted transcripts (blue) identified by expression meta-analysis. (B) Quantification of the change in translation efficiency in miR-203 KO samples for miR-203 targets identified by expression meta-analysis. (C) Comparison of translation efficiency for miR-203 targets identified by Ago2-HITS-CLIP (red) and non-targeted transcripts (blue). (D) Quantification of the change in translation efficiency in miR-203 KO samples for miR-203 targets based on Ago2-HITS-CLIP.
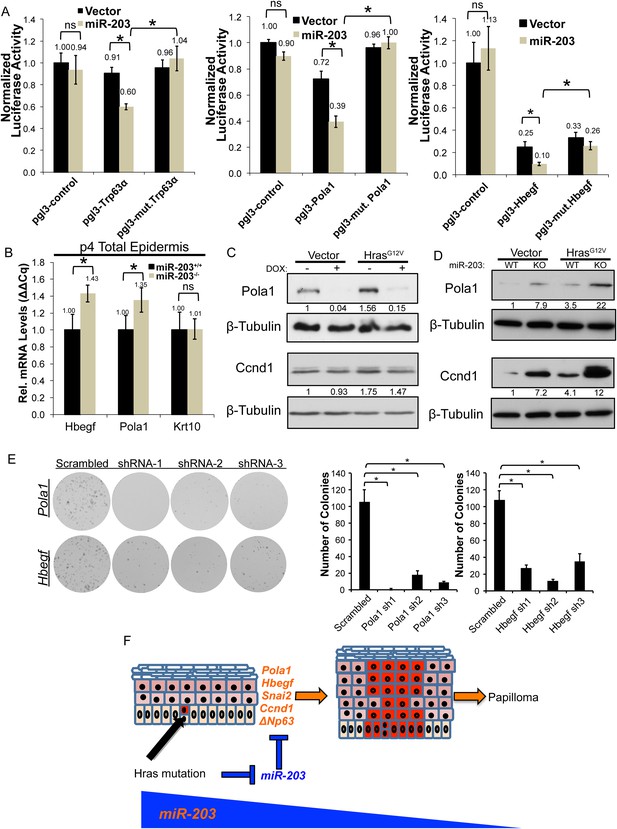
Hbegf and Pola1 are direct miR-203 target genes critical for keratinocyte proliferation.
(A) 3′UTR luciferase reporter assays demonstrate that miR-203 directly targets Trp63 (positive control), Pola1, and Hbegf in keratinocytes (representative of n = 3 independent experiments, mean ± propagated standard deviation displayed, *p < 0.05, ns = non-significant, Student's t-test two-sided) (B) Hbegf and Pola1 are upregulated in miR-203−/− isolated epidermis (p4) (n = 8 and n = 10, miR-203+/+ and miR-203−/− animals respectively, mean ± SEM displayed, *p < 0.05). (C, D) Western blots from lysates with miR-203 overexpression (48 hr) or miR-203 ablation. (E) shRNA knockdown of Hbegf or Pola1 impairs keratinocyte colony formation ability (representative of n = 3 independent experiments, *p <0.05, mean ± standard deviation displayed). (F) Model for the mechanism of miR-203 in restricting Hras-initiated tumorigenesis.
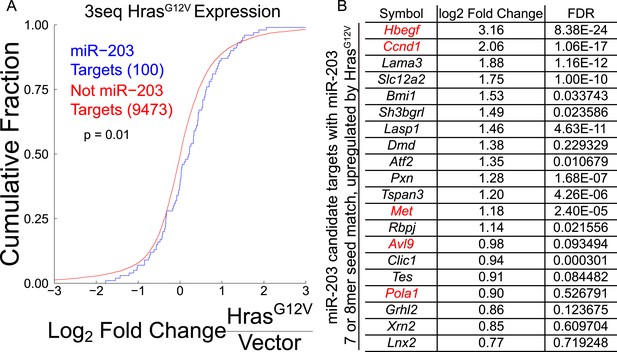
A subset of miR-203 targets are upregulated by HrasG12V.
(A) miR-203 target genes identified through meta-analysis containing miR-203 seed matches are more likely to be upregulated upon HrasG12V expression in primary keratinocytes than non-targeted transcripts (p-value ≤0.05, K–S test one-sided). (B) Top 20 miR-203 targets, based on expression meta-analysis, ranked by upregulation by HrasG12V, genes shown in red are also identified by Ago2-HITS-CLIP.
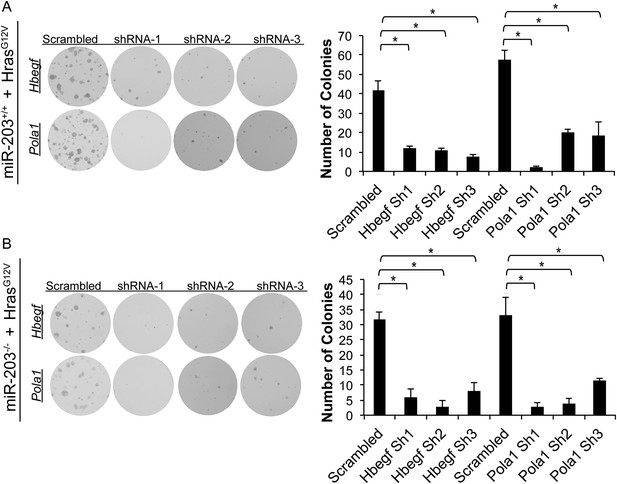
Hbegf and Pola1 are required for keratinocyte growth potential in HrasG12V-transformed miR-203+/+ and miR-203−/− cultures.
(A) Colony formation assays of established miR-203+/+ cultures stably infected with pbabe-HrasG12V-neo, followed by Pola1 and Hbegf knockdown. (B) Colony formation assays of established miR-203+/+ cultures stably infected with pbabe-HrasG12V-neo, followed by Pola1 and Hbegf knockdown. (p-value determined by ANOVA with Tukey HSD correction, n = 3 wells per experiment).
Additional files
-
Supplementary file 1
Sequencing mapping statistics.
- https://doi.org/10.7554/eLife.07004.023
-
Supplementary file 2
Primers, antibodies, and shRNAs used in this study.
- https://doi.org/10.7554/eLife.07004.024





