A panel of induced pluripotent stem cells from chimpanzees: a resource for comparative functional genomics
Figures
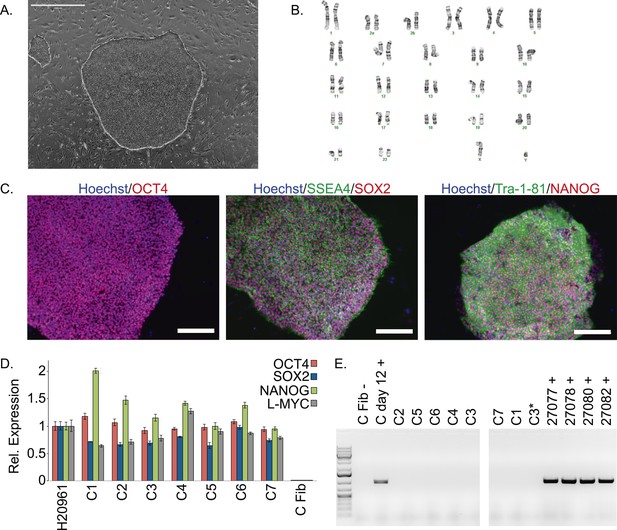
Characterization of chimpanzee induced pluripotent stem cell (iPSC) lines.
(A) Phase contrast image of representative chimpanzee iPSC line. Scale bar: 1000 μm. (B) Representative karyotype from chimpanzee iPSC line after >15 passages, showing no abnormalities. (C) ICC staining of iPSC lines with antibodies for pluripotency markers as indicated. Scale bar: 200 μm. (D) Quantitative PCR testing for expression of endogenous pluripotency factors in all 7 chimpanzee iPSC lines. Line H20961 is a male human iPSC line generated in-house used as reference. (E) PCR gel showing an absence of exogenous episomal reprogramming factors in all 7 chimpanzee iPSC lines. All PCRs were carried out on templates extracted from passage >15 with the exception of C3651*, which is from passage 2. Fib—is a negative fibroblast control (from individual C8861) prior to transfection, day 12 + is a positive control 12 days after transfection, 27,077 + to 27,082 + are the plasmids used for reprogramming.
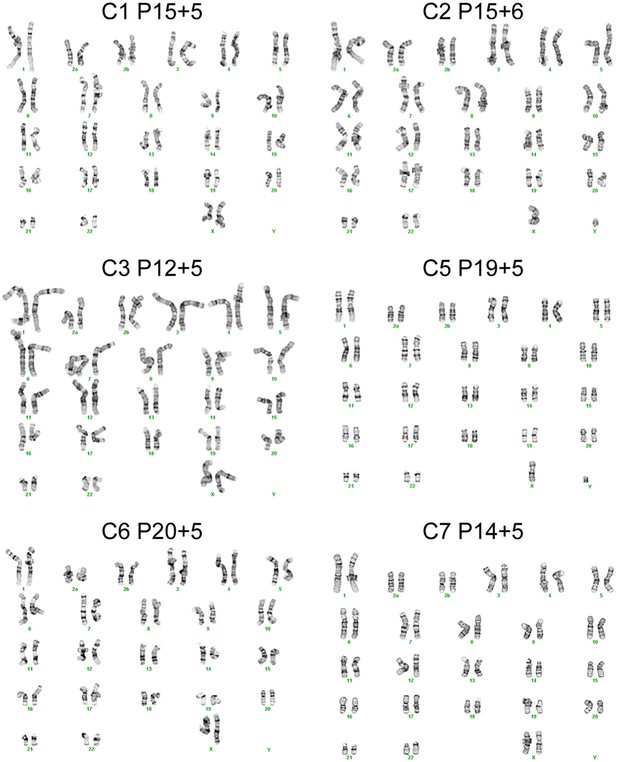
Karyotypes for the 6 chimpanzee iPSC lines not shown in main text figures, generated after >15 passages in culture.
Passage number for each line represents passages on mouse embryonic fibroblast (MEF) feeders plus additional passages on Matrigel.
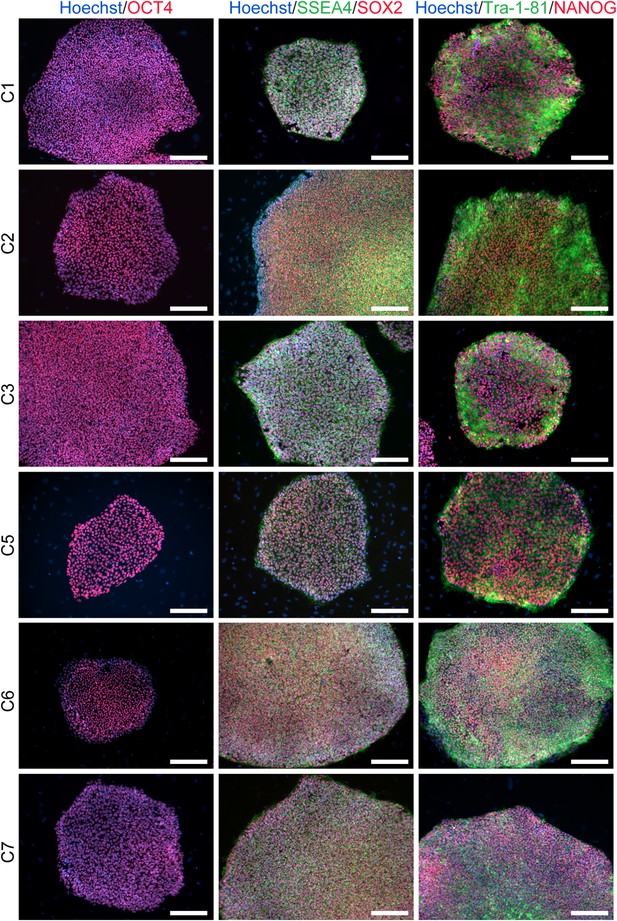
ICC staining of the 6 chimpanzee iPSC lines not shown in main text figures with antibodies for pluripotency markers as indicated.
Scale bar: 200 μm.
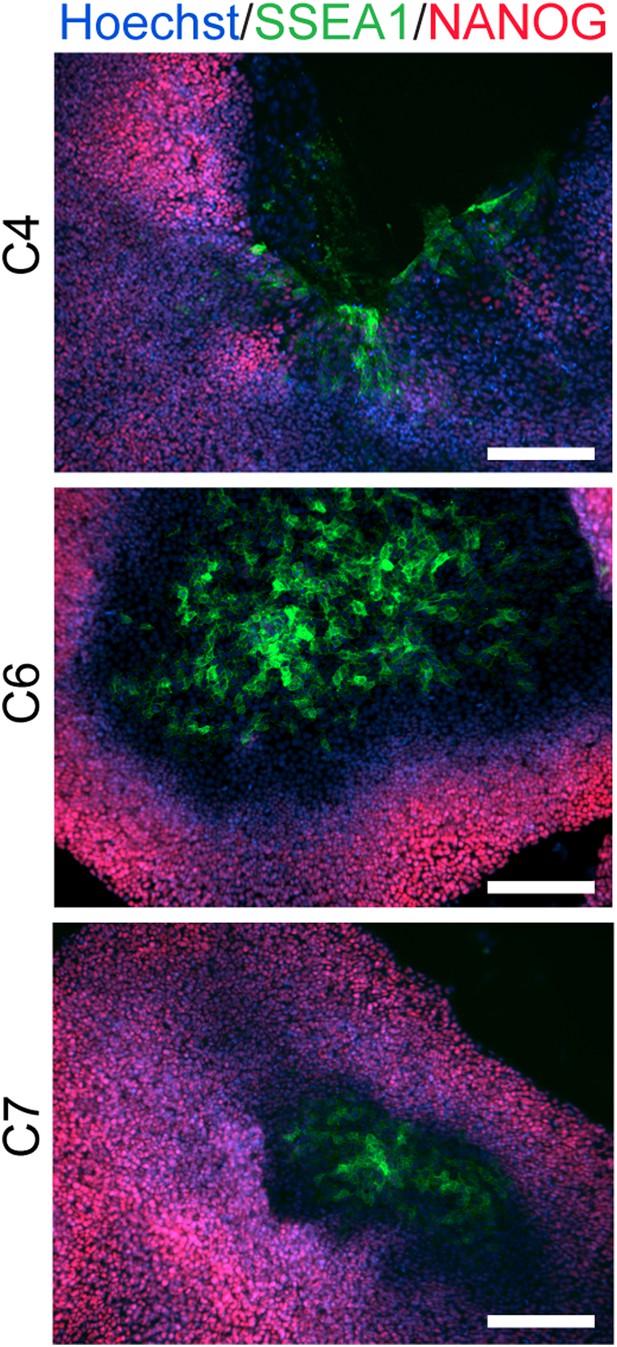
ICC staining showing SSEA1 expression in chimpanzee iPSC culture plates, clearly distinct from NANOG expression.
https://doi.org/10.7554/eLife.07103.006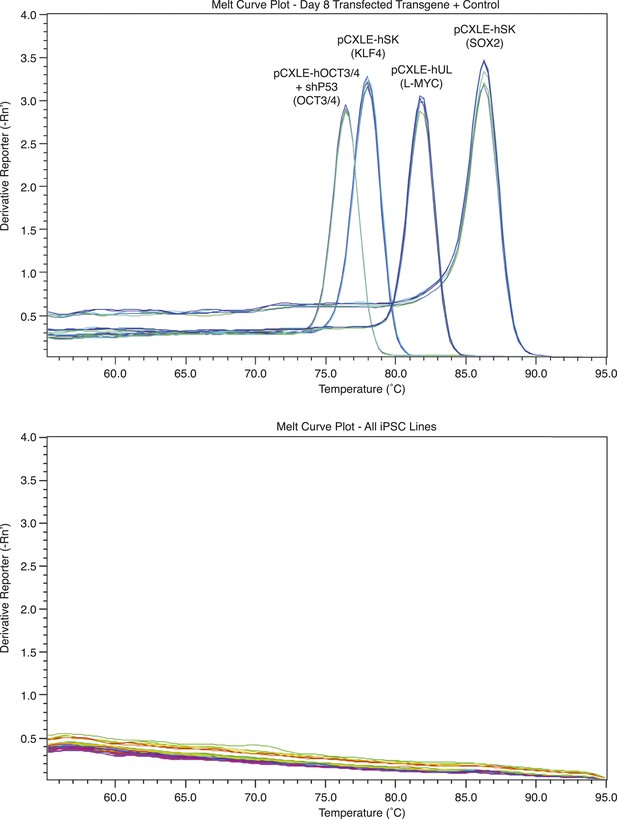
Melt curves showing a lack of exogenous reprogramming gene expression in episomally reprogrammed chimpanzee iPSCs after >10 passages.
https://doi.org/10.7554/eLife.07103.007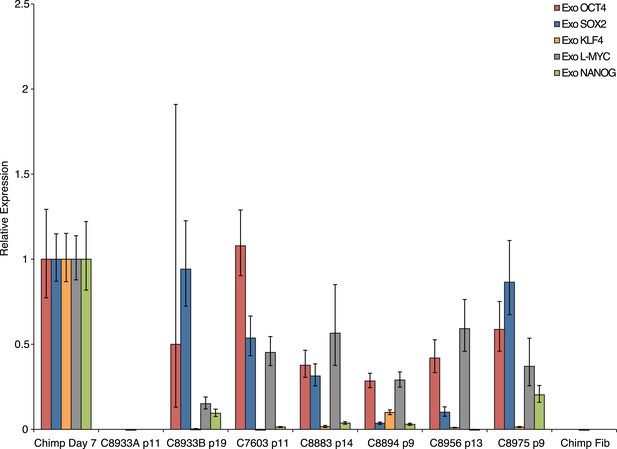
Exogenous gene expression in retrovirally reprogrammed chimpanzee iPSCs after various passages.
All values are relative to expression in a day-7-post-transfection chimpanzee fibroblast.
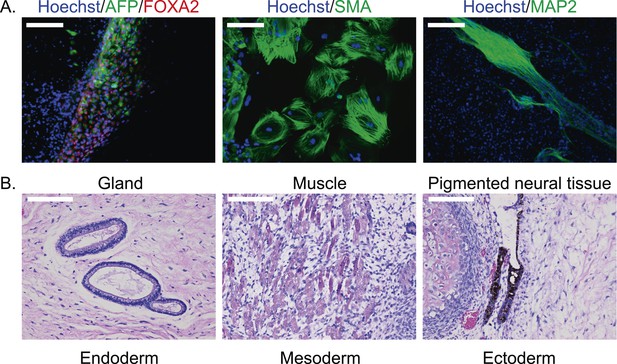
(A) ICC staining of differentiated embryoid bodies with antibodies for the three germ layers as indicated.
Scale bar: 200 μm. (B) Histological staining of teratomas derived from iPSC line C4955, showing generation of tissues from all three germ layers. Scale bar: 100 μm.
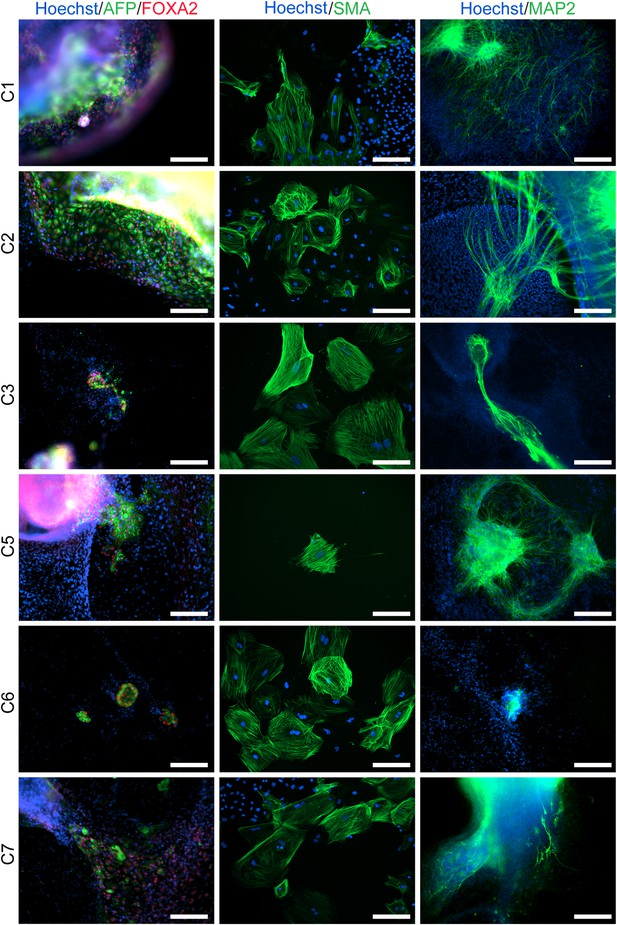
ICC staining of differentiated embryoid bodies derived from the 6 chimpanzee iPSC lines not shown in main text figures, with antibodies for the three germ layers as indicated.
Scale bar: 200 μm.

ICC staining of directly differentiated hepatocytes from line C2, with antibodies as indicated.
Scale bar: 200 μm.
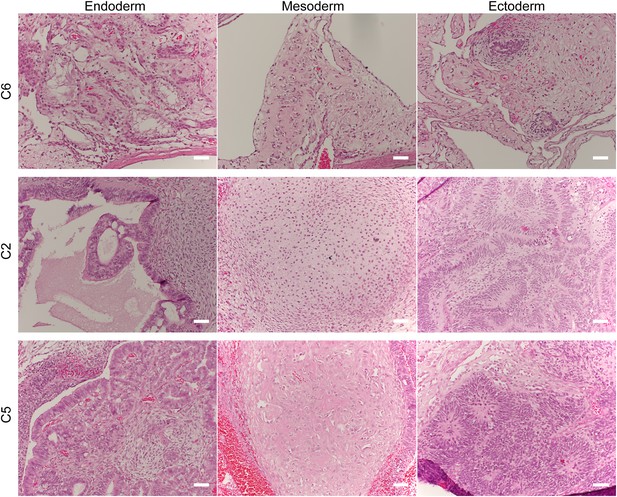
Histological staining of teratomas derived from three additional chimpanzee iPSC lines, showing generation of tissues from all three germ layers.
Scale bar: 500 μm.
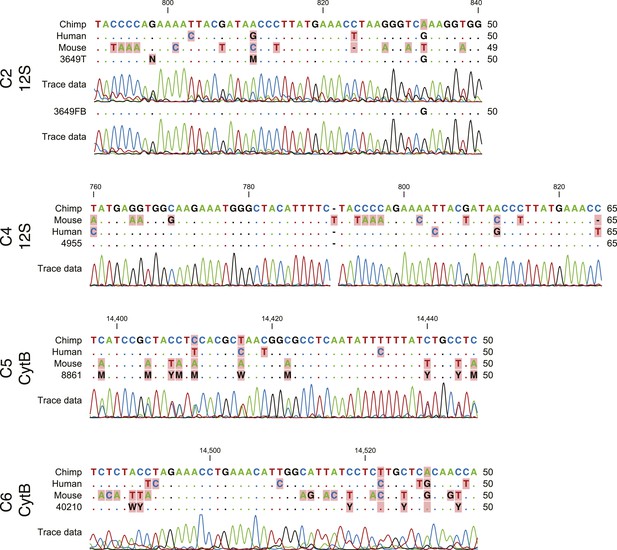
Sequencing traces from teratomas generated from chimpanzee iPSC lines for the mitochondrial genes 12S (C3649, C4955) and cytb (C8861, C40210).
All traces show clear evidence of the presence of chimpanzee tissue in the teratoma.
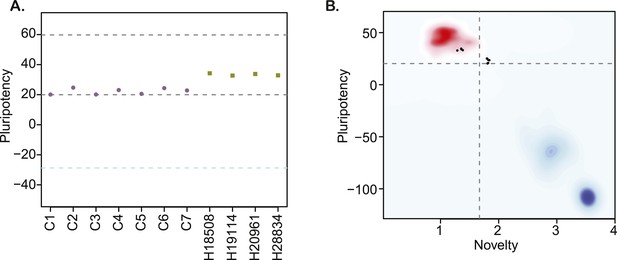
(A) PluriTest pluripotency scores in the 7 chimpanzee lines and 4 human reference iPSC lines.
Purple circles denote chimpanzees; yellow squares, humans. (B) PluriTest results after removal of probes not mapping to the chimpanzee genome. All samples in the top left quadrant are human and have satisfactory pluripotency and novelty scores. Samples in the top right quadrant correspond to our chimpanzee iPSC panel, and have consistently high pluripotency yet high novelty scores.
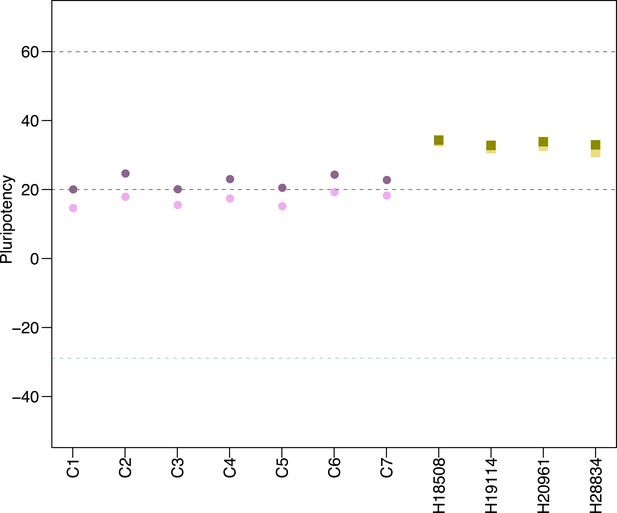
The effects of probe sub-setting in PluriTest pluripotency score calculations.
Lighter shades indicate pluripotency scores before the removal of probes not mapping to the chimpanzee genome, darker shades indicate pluripotency after probe removal. Purple circles denote chimpanzees; yellow squares, humans.
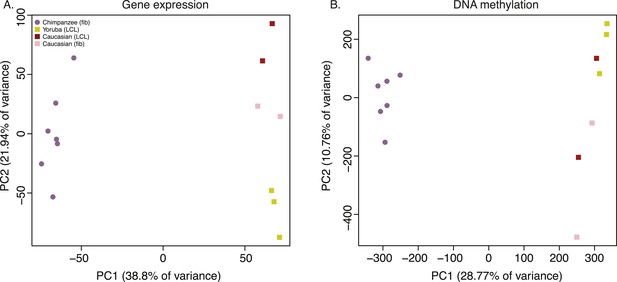
Principal component (PC) analysis plots of data from the iPSCs.
(A) Principal component analysis (PCA) generated from expression data of 12,171 orthologous genes. (B) PCA generated from DNA methylation data measured by 335,307 filtered probes.
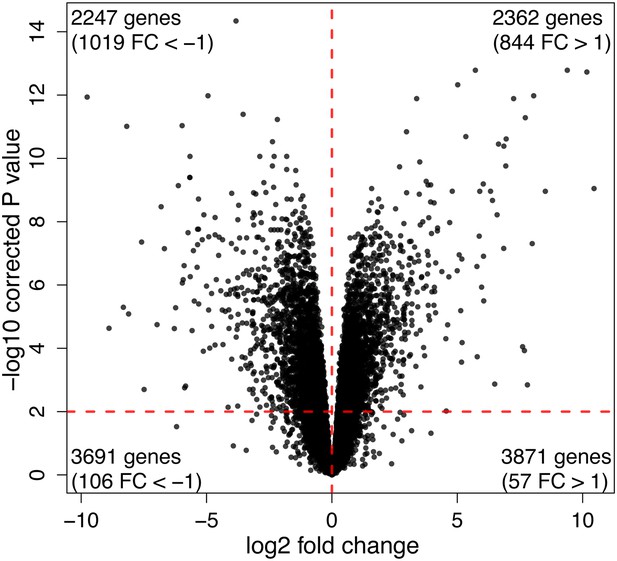
Volcano plot showing the distribution of DE genes between iPSCs of chimpanzee and human origin.
https://doi.org/10.7554/eLife.07103.018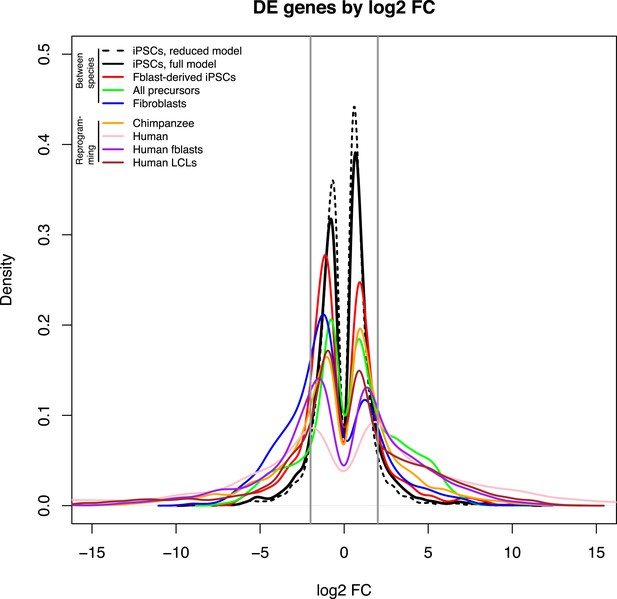
Density plots of log2 FC change values amongst DE genes for the main comparisons presented in the text.
The area bounded by the grey lines represents log2 FC changes with an absolute magnitude <2.
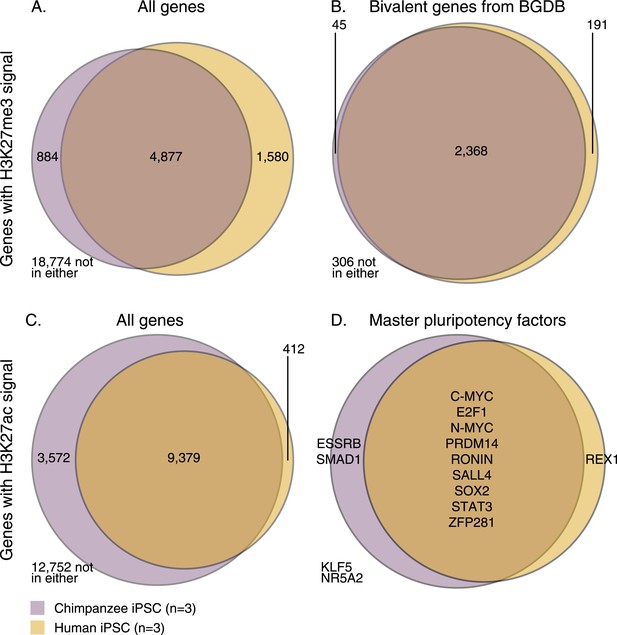
Overlap of H3K27me3 and H3K27ac signal between chimpanzee and human iPSCs at orthologous transcription start sites (TSSs).
(A) H3K27me3 enrichment near all genes with an orthologous TSS. (B) H3K27me3 enrichment near 2910 genes previously identified as bivalent in human PSCs. (C) H3K27ac enrichment near all genes with an orthologous TSS. (D) H3K27ac peaks near 14 known pluripotency master regulators with orthologous TSSs.
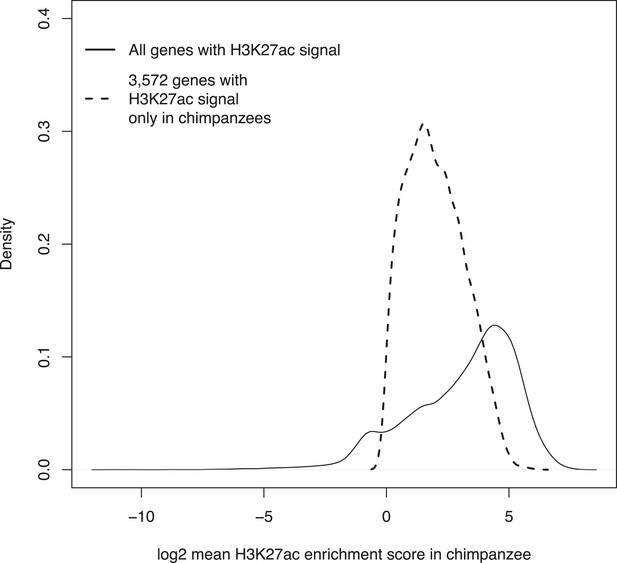
Density plots of H3K27ac enrichment scores at orthologous TSSs in the entire data set and at 3572 genes enriched only in chimpanzee iPSCs.
https://doi.org/10.7554/eLife.07103.021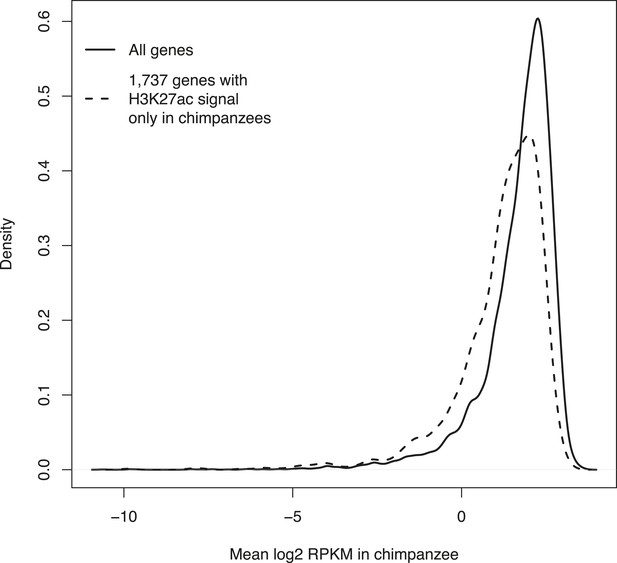
Density plots of mean RPKM in chimpanzee iPSCs in all 12,171 genes with expression data and in the subset of 1737 genes with expression data and H3K27ac signal enrichment solely in chimpanzee iPSCs.
https://doi.org/10.7554/eLife.07103.022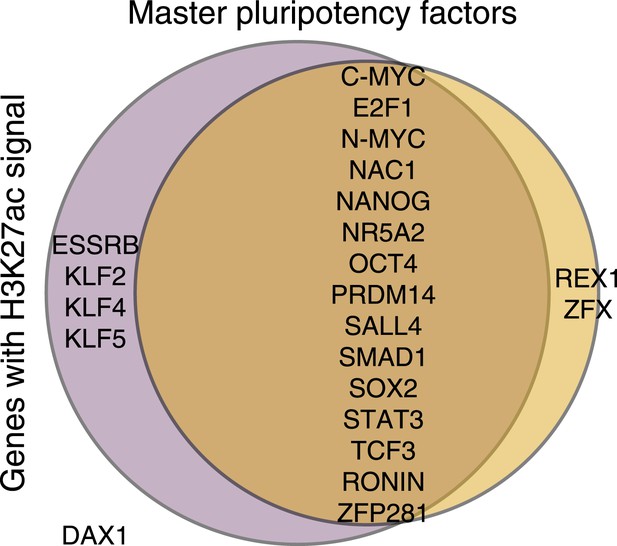
H3K27ac peaks observed in at least 1 chimpanzee or human iPSC, as identified by MACS at 22 known pluripotency master regulators.
In the case of all three genes that differ between this figure and Figure 5D—KLF5, NR5AD and SMAD1—processed enrichment signal after accounting for orthology is weak, and falls very close to our normalised enrichment score threshold of 1, explaining the difference between the two.
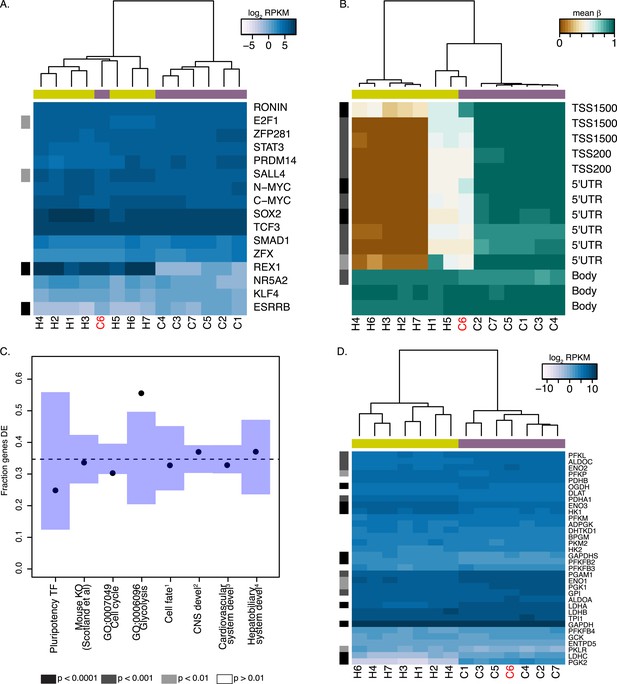
REX1 may be dispensable for pluripotency in chimpanzee iPSCs.
In both panels REX1-expressing chimpanzee iPSC line is coloured red, significant interspecies differences are indicated along the left-hand side, and purple boxes indicate chimpanzee lines, yellow boxes indicate human lines. (A) Expression values of 16 core pluripotency transcription factors in all human and chimpanzee iPSC lines. (B) Methylation status of 13 CpG sites associated with REX1 in all human and chimpanzee iPSCs. Location of the probe relative to the gene sequence is indicated along the right hand side. (C) Fraction of differentially expressed (DE) genes in multiple categories downstream of REX1 in human and mouse ESCs. 1: Genes associated with any Gene Ontology term that contains the words ‘ectoderm’, ‘mesoderm’ or ‘endoderm’. 2: CNS development genes are associated with GO:0007417 or any of its offspring. 3: cardiovascular system development genes are associated with GO:0072358 or any of its offspring. 4: hepatobiliary system development genes are associated with GO:0055123 or any of its offspring. (D) Expression levels of 34 genes associated with GO:0006096, glycolysis, in all human and chimpanzee iPSC lines. All reported p-values were calculated after excluding C6.
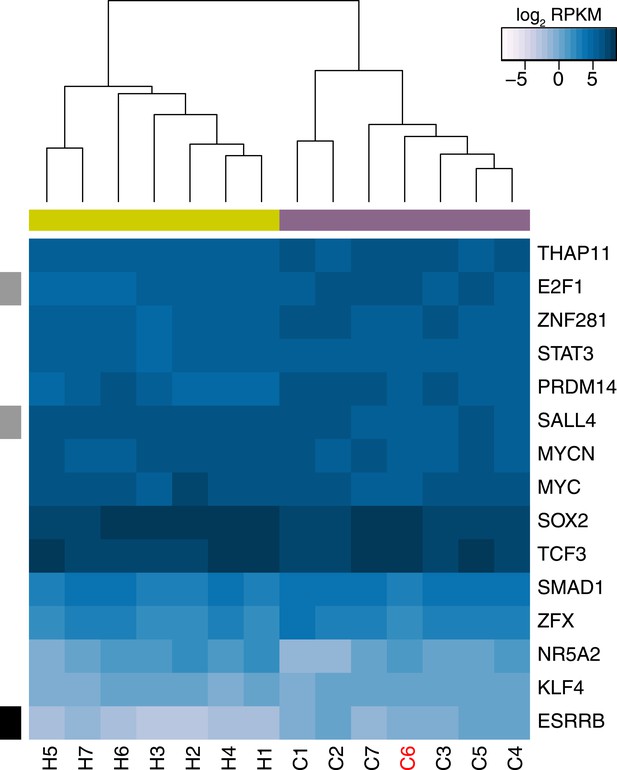
Expression values of 15 core pluripotency transcription factors in all human and chimpanzee iPSC lines.
The data used to generate this figure are identical to those used to generate Figure 5A except that expression levels of REX1 are not included in the calculation. REX1-expressing chimpanzee iPSC line C6 is coloured red, significant interspecies differences are indicated along the left-hand side, and purple boxes indicate chimpanzee lines, yellow boxes indicate human lines.
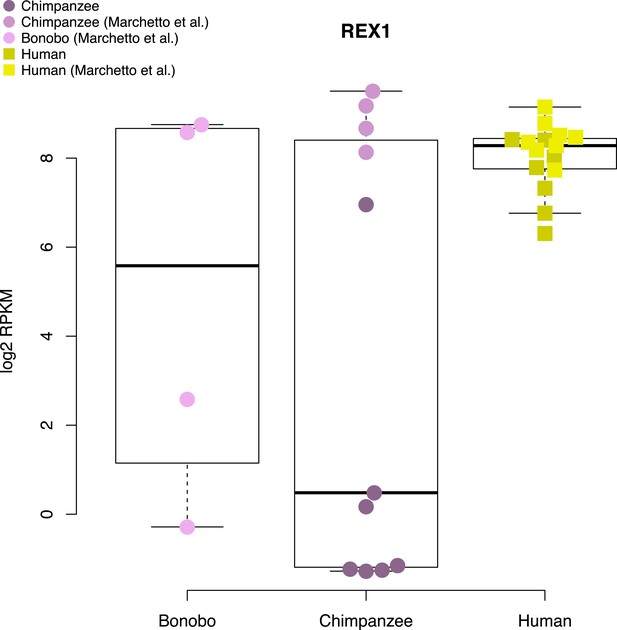
Expression levels of REX1 in human, chimpanzee and bonobo iPSC lines generated in this study and in Marchetto et al. (2013b).
Data for this figure were jointly normalised.
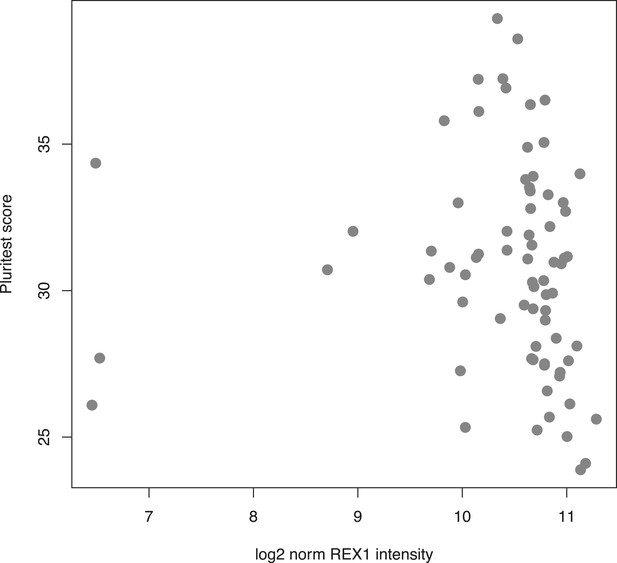
Plot of PluriTest pluripotency scores vs normalised REX1 intensity in 73 human iPSC lines derived in-house.
https://doi.org/10.7554/eLife.07103.027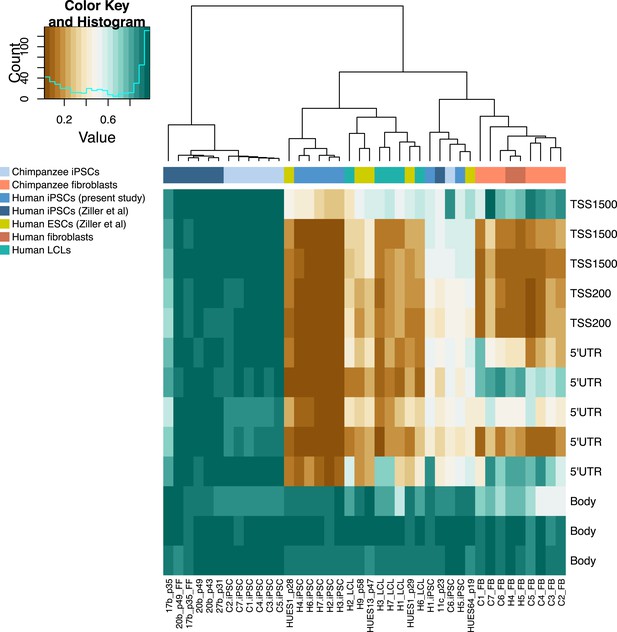
Methylation status of 13 CpG sites associated with REX1 in chimpanzee and human iPSCs from this study and human PSCs from Ziller et al. (2011).
REX1-expressing chimpanzee iPSC line is coloured red; location of the probes relative to the gene sequence is indicated along the right hand side.
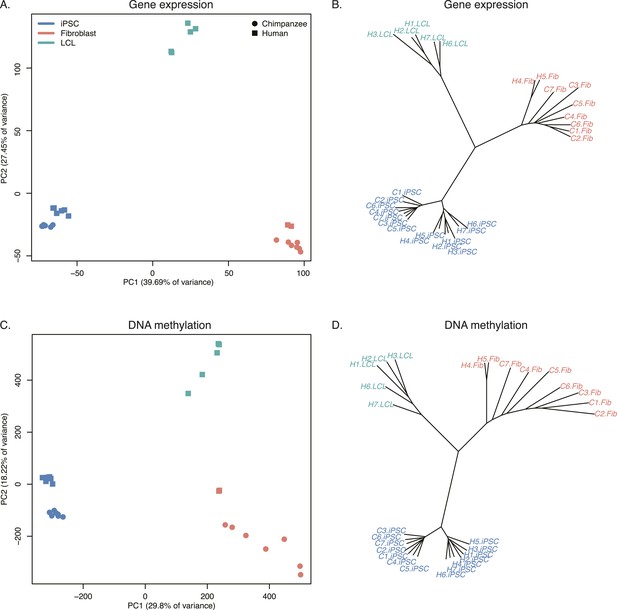
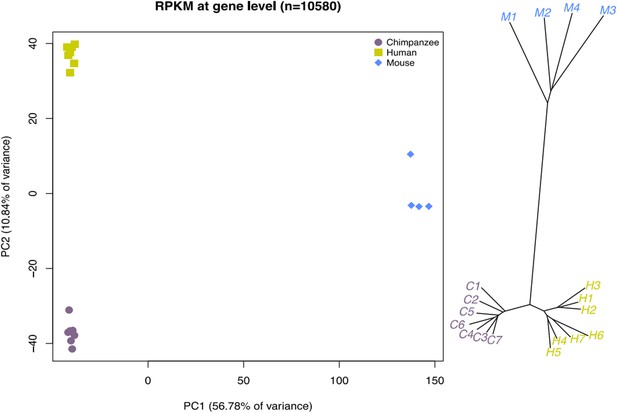
Relationships of iPSCs to their precursors.
(A) PCA of gene expression data from all iPSCs and their precursor cell lines. (B) Neighbour-joining tree of Euclidean distances between all samples generated based on the gene expression data. (C) PCA of DNA methylation data from all iPSCs and their precursor cell lines. (D) Neighbour-joining tree of Euclidean distances between all samples generated based on the DNA methylation data.
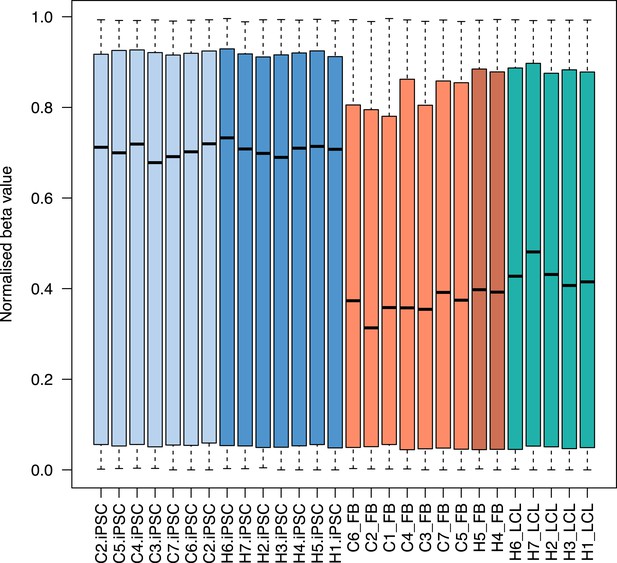
Boxplots of methylation beta values at 335,307 probes across all samples.
Plots are colored by tissue type: light blue: chimpanzee iPSCs; dark blue: human iPSCs; light orange: chimpanzee fibroblasts; dark orange: human fibroblasts; turquoise: human lymphoblastoid cell lines (LCLs).
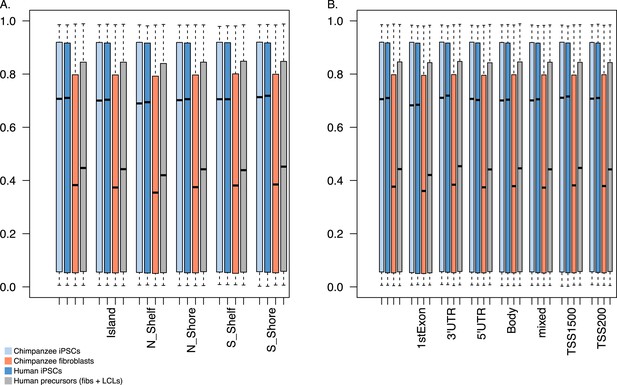
Boxplots of methylation beta values across all samples, grouped by potency and genomic features.
Boxes are colored by tissue type: light blue: chimpanzee iPSCs; light orange: chimpanzee fibroblasts. (A) By methylation feature; (B) By relative position.
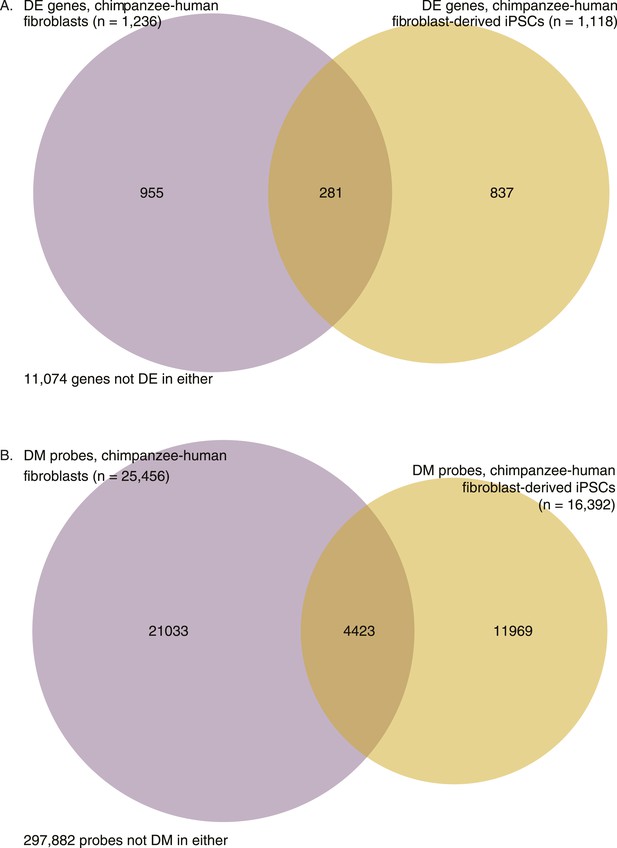
Venn diagrams showing overlap in interspecies differences before and after reprogramming.
(A) Overlap in DE genes between chimpanzee and human fibroblasts, and chimpanzee and human fibroblast-derived iPSCs. (B) Overlap in differentially methylated (DM) probes between chimpanzee and human fibroblasts, and chimpanzee and human fibroblast-derived iPSCs.
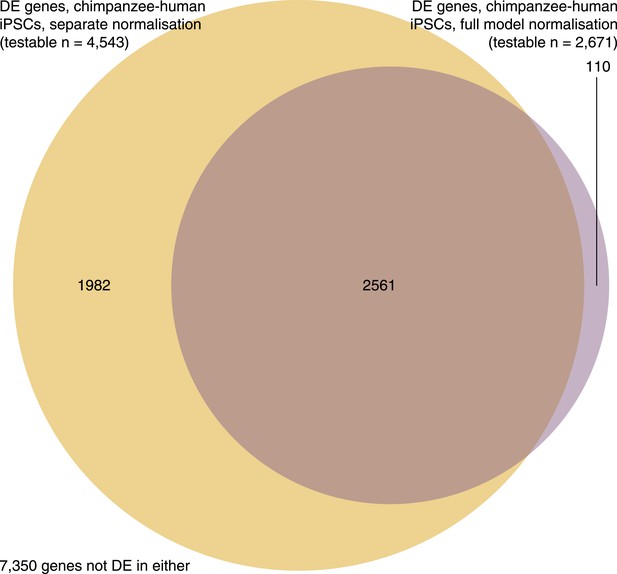
Venn diagram showing overlap of genes identified as DE between iPSCs of the two species when we normalize the iPSC data independently and alongside data from the precursors.
https://doi.org/10.7554/eLife.07103.033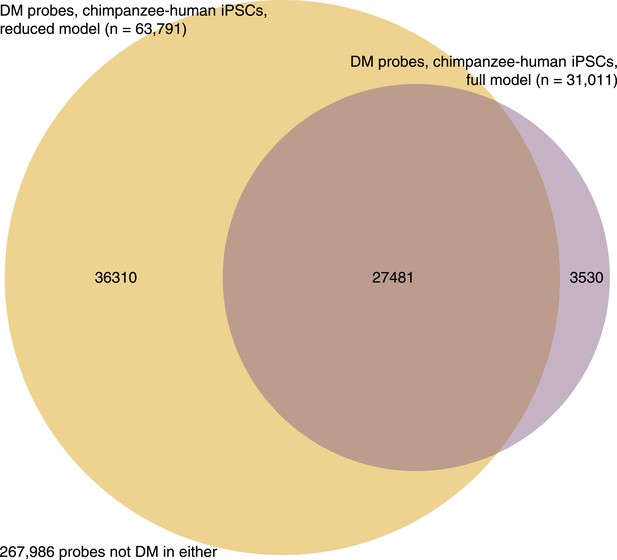
Venn diagram showing overlap of probes identified as DM between iPSCs of the two species under the full and reduced limma models.
https://doi.org/10.7554/eLife.07103.034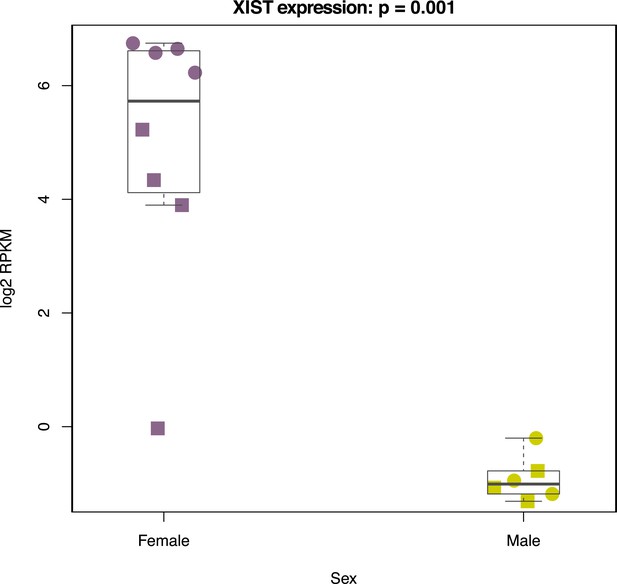
Normalized XIST expression values in 7 chimpanzee and human iPSCs.
Circles denote chimpanzee iPSCs, squares indicate human iPSCS.
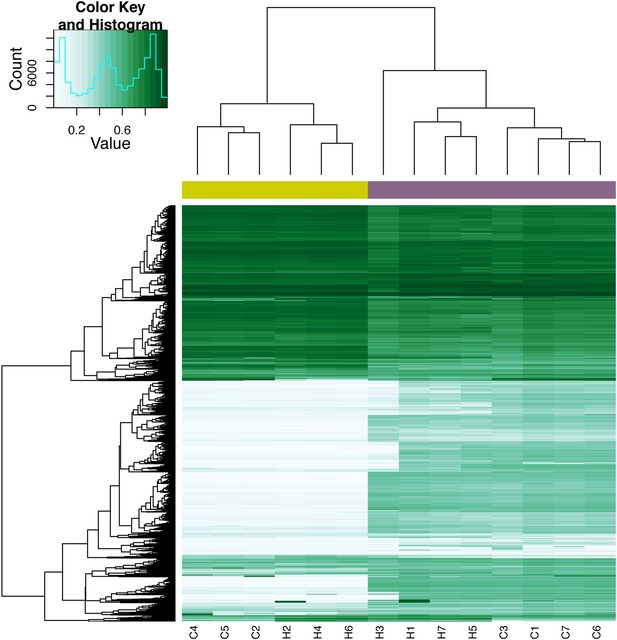
Quantile-normalized methylation beta values at 8210 X-chromosome probes in 7 chimpanzee iPSCs and 7 human iPSCs.
The colour bar beneath the dendrogram indicates sex of the individuals: purple: female; yellow: male. Sample names ending with _FB indicate fibroblast lines used to generate the corresponding iPSC line, samples ending with _LCL indicate LCL lines used to generate the corresponding iPSC line.
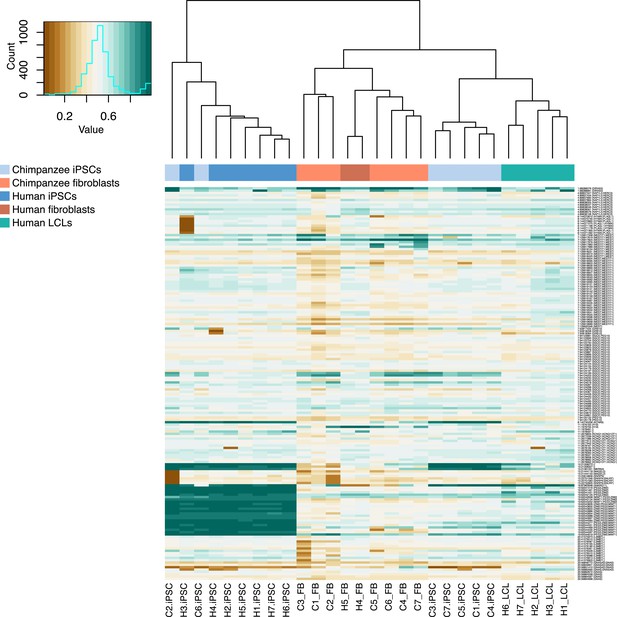
Normalized methylation beta values at 168 assayable probes known to be subject to parental imprinting effects, from Ma et al. (2014).
Sample names ending with _FB indicate fibroblast lines used to generate the corresponding iPSC line, samples ending with _LCL indicate LCL lines used to generate the corresponding iPSC line.
Videos
Calcium transient flux in and out (GFP labelled) and contractility of directly differentiated cardiomyocytes from chimpanzee iPSC line C7.
https://doi.org/10.7554/eLife.07103.014Additional files
-
Supplementary file 1
Description of samples. a: Descriptive data for all chimpanzee cell lines used in this work. b: Descriptive data for all human iPSC lines used.
- https://doi.org/10.7554/eLife.07103.038
-
Supplementary file 2
Origin and purpose of all primers used.
- https://doi.org/10.7554/eLife.07103.039
-
Supplementary file 3
Normalized RPKM values and DE genes between chimpanzee and human iPSCs.
- https://doi.org/10.7554/eLife.07103.040
-
Supplementary file 4
Gene Ontology BP terms associated with genes DE between chimpanzee and human iPSCs.
- https://doi.org/10.7554/eLife.07103.041
-
Supplementary file 5
DMRs identified between chimpanzee and human iPSCs.
- https://doi.org/10.7554/eLife.07103.042
-
Supplementary file 6
Genome-wide data summary statistics. a: Numbers of DM probes and DMRs between chimpanzee and human iPSCs identified under various mean β difference thresholds. b: RNA- and ChIP-sequencing reads generated and mapped for all samples in this work. c: Effects of different normalization schemes on the number of genes classified as DE in the full data set.
- https://doi.org/10.7554/eLife.07103.043
-
Supplementary file 7
Gene Ontology BP terms associated with genes within DMRs between chimpanzee and human iPSCS.
- https://doi.org/10.7554/eLife.07103.044
-
Supplementary file 8
H3K27ac and H3K27me3 enrichment scores in 3 chimpanzee and 3 human iPSCs around 26,115 orthologous TSSs.
- https://doi.org/10.7554/eLife.07103.045
-
Supplementary file 9
Correlations between principal components and selected covariates. a: in the expression data. b: in the methylation data.
- https://doi.org/10.7554/eLife.07103.046
-
Supplementary file 10
Normalized RPKM values and DE genes identified under the full limma DE testing framework.
- https://doi.org/10.7554/eLife.07103.047
-
Supplementary file 11
DMRs identified between chimpanzee iPSCs and their precursor fibroblasts.
- https://doi.org/10.7554/eLife.07103.048
-
Supplementary file 12
DMRs identified between human iPSCs and their precursor cells.
- https://doi.org/10.7554/eLife.07103.049





