Receptor tyrosine kinases modulate distinct transcriptional programs by differential usage of intracellular pathways
Figures
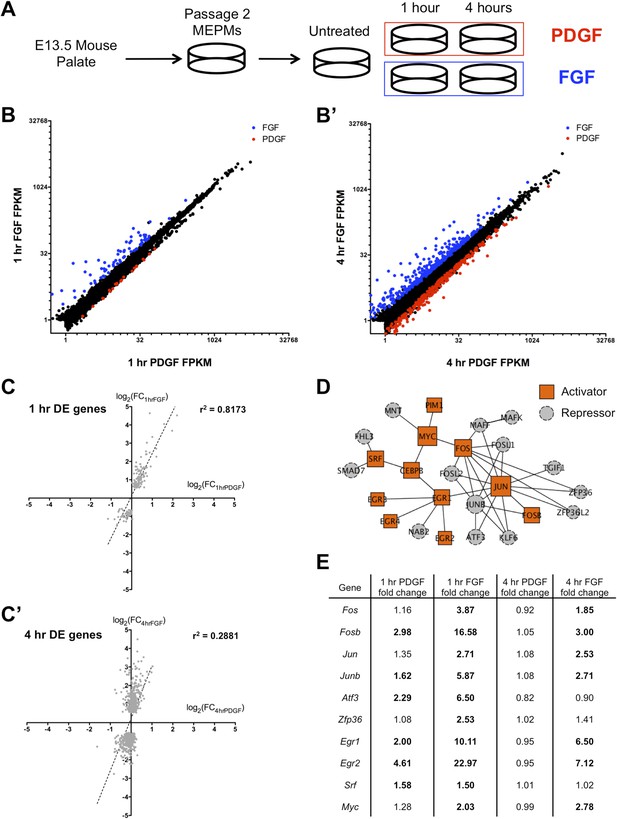
FGF and PDGF stimulation result in distinct transcriptional responses.
(A) Mouse embryonic palatal mesenchyme (MEPM) cells were dissected from E13.5 embryos, passaged twice, and then serum starved overnight prior to stimulation with either PDGFAA or FGF1 + heparin. (B) Expression of all genes with FPKM (fragments per kilobase of exon per million reads mapped) >1 at 1 hr (11,217 genes) and (B') 4 hr (11,266 genes). Genes colored blue are significantly increased with fibroblast growth factor (FGF) treatment and genes colored red are significantly increased with platelet-derived growth factor (PDGF) treatment. Values plotted on log2 scale. (C) Fold change (FC) comparison for all differentially expressed (DE) genes at (C) 1 hr or (C') 4 hr (compared to serum starved sample) shows high correlation between the transcriptional response to each growth factor at 1 hr but low correlation at 4 hr. (D) Protein–protein interaction (PPI) network for all genes upregulated at 1 hr contains many classic immediate early genes (such as AP-1 components, Myc, and Srf). Genes are colored based on their primary reported role in transcriptional regulation (22, 23), with orange squares representing activators and gray circles representing repressors. (E) FC (compared to untreated sample) for selected genes upregulated at 1 hr. Genes in bold are induced >1.5-fold in response to the indicated growth factor. Although both PDGF and FGF regulate many shared targets, the induction in response to FGF exhibits greater magnitude (Fos, Fosb, Junb, Atf3, Egr1, Egr2) and longer duration (Fos, Fosb, Jun, Junb, Egr1, Egr2).
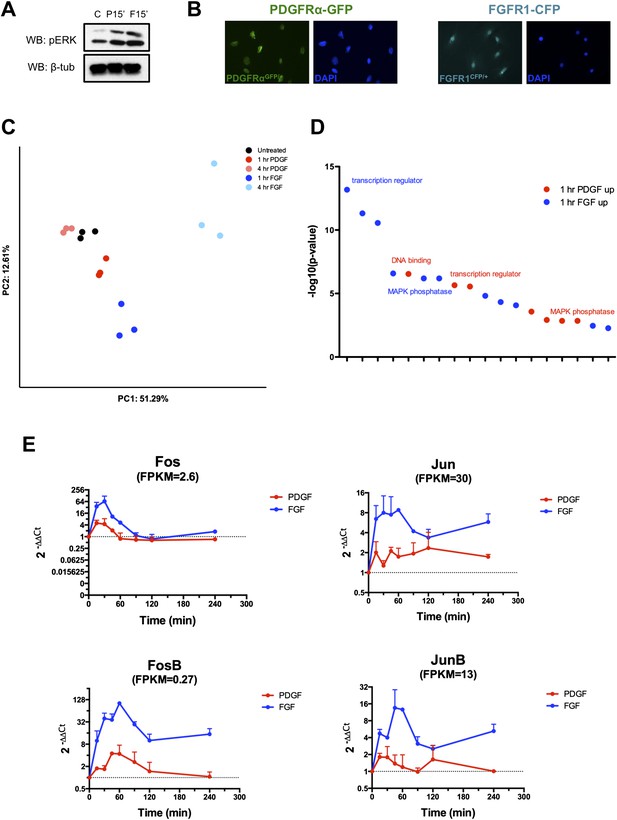
Characterization of E13.5 MEPMs and transcriptional response to PDGF and FGF signaling.
(A) In the samples submitted for RNA-seq, both PDGF and FGF induce robust pERK responses at 15 minutes. (B) MEPMs derived from either Pdgfra-GFP or Fgfr1-CFP knockin reporter embryos express each receptor in all cells, confirming MEPMs are a homogenous cell population at the level of PDGF and FGF receptor expression. (C) Principal component analysis (PCA) reveals distinct responses to PDGF and FGF signaling, with the first principal component (51.29% of variance) segregating conditions based on ligand treatment and the second principal component (12.61% of variance) separating samples based on duration of stimulation. (D) Gene ontology analysis (molecular function) on genes induced at 1 hr reveals an overrepresentation of transcription factors and MAP kinase phosphatases (DUSPs). (E) Validation of selected transcription factors by qPCR confirms greater magnitude and duration of induction by FGF compared to PDGF.
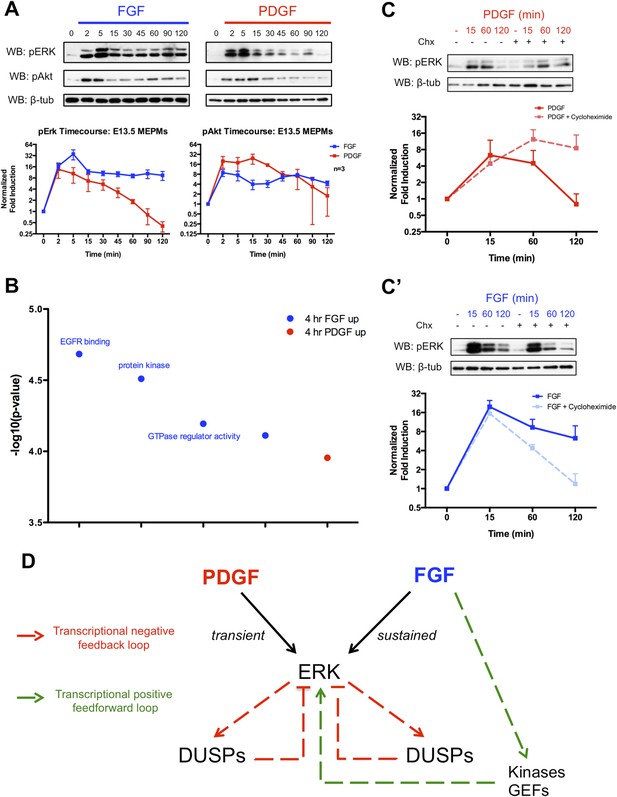
The delayed transcriptional response provides differential regulation of pERK duration in response to FGF and PDGF signaling.
(A) Signaling time course shows a more robust phospho-ERK (pERK) response to FGF (blue) than PDGF (red) and a similar pAkt response to both growth factors. (B) Gene ontology analysis (molecular function) of genes DE between the 4-hr PDGF and 4-hr FGF conditions indicates enrichment for kinases and GTPase regulators in response to FGF signaling. (C) Cycloheximide treatment has opposite effects on pERK duration following (C) PDGF and (C') FGF stimulation, indicating the delayed transcriptional response (dependent on protein synthesis and thus inhibited by cycloheximide) can provide both negative and positive signals to modulate pERK kinetics. (D) Model depicting loops that regulate the duration of the pERK wave in response to receptor tyrosine kinase (RTK) signaling includes both negative (dual-specificity phosphatases [DUSPs]) and positive (kinases, GEFs) components from the delayed transcriptional response. Western blot quantification plotted as mean ± SEM, n = 3.
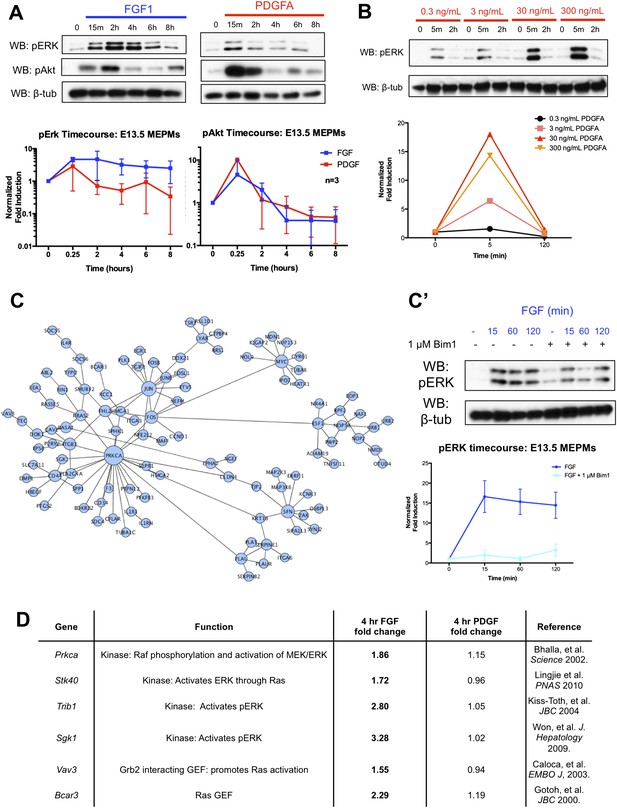
Signaling kinetics and organization of the late transcriptional response in response to PDGF and FGF treatment.
(A) Long timecourse of PDGF and FGF treatment followed by Western blot for pERK and pAkt shows the FGF mediated pERK response persists up to 6 hours following growth factor stimulation. (B) The duration of the pERK response to PDGFA treatment does not change with increasing ligand dosage, indicating 30 ng/mL is a saturating dose for pERK duration. (C) PPI network constructed from genes differentially expressed at 4 hours following FGF treatment reveals Prkca as a highly connected node, suggesting Protein Kinase C as a regulator of the pERK driven FGF response. (C') Inhibition of PKC abolishes both the initial pulse and sustained wave of pERK in response to FGF treatment. (D) Many candidate kinases and GEFs, such as Prkca (Bhalla et al., 2002), Stk40 (Li et al., 2010), Trib1 (Kiss-Toth et al., 2004), Sgk1 (Won et al., 2009), Vav3 (Caloca et al., 2003), and Bcar3 (Gotoh, et al. 2000), have reported roles in positively modifying MEK/ERK signaling. Prkca
FGF and PDGF transcriptional responses exhibit differential usage of intracellular pathways.
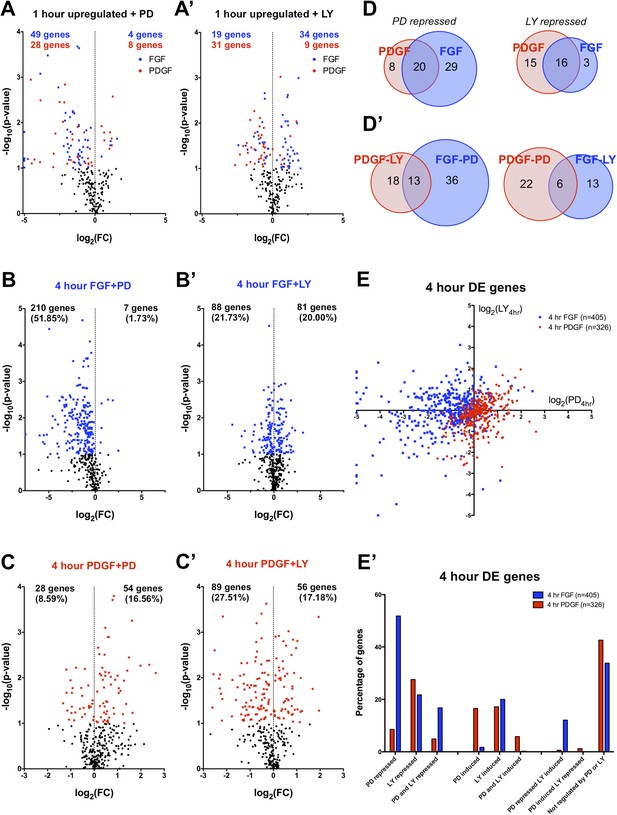
(A) Volcano plots visualizing the effect of (A) MEK (Mitogen/Extracellular signal-regulated kinase) inhibition and (A') phosphatidylinositol 3-kinase (PI3K) inhibition on the shared set of 113 genes upregulated at 1 hr reveal that FGF (blue points) shows increased dependence on MEK/extracellular signal-related kinase (ERK) signaling, while PDGF (red points) utilizes PI3K to a greater degree. X-axis plotted as log2([1 hr ligand + inhibitor]/[1 hr ligand]). (B) Genes with increased expression at 4-hr FGF treatment show higher dependence on (B) MEK/ERK activity compared to (B') PI3K. (C) In contrast, genes with increased expression at 4-hr PDGF treatment show greater usage of (C') PI3K compared to (C) MEK/ERK. X-axis plotted as log2([4 hr ligand + inhibitor]/[4 hr ligand]). Data analyzed using two sample t-test, and genes at p < 0.1 are colored significant. Black points represent genes not significant at this threshold in all plots. (D) A core set of MEK/ERK (20 genes) and PI3K (16 genes) are dependent on these pathways downstream of both PDGF and FGF. (D') A minority of genes can be activated through either MEK/ERK or PI3K signaling in response to PDGF or FGF, indicating a degree of plasticity in intracellular pathway usage. (E) Scatter plot comparing effect of MEK/ERK and PI3K inhibition on all 4 hr DE genes reflects FGF-ERK and PDGF-PI3K dependencies. Data plotted as log2([4 hr ligand + inhibitor]/[4 hr ligand]) and capped at ±5 for visualization. (E') 52% of FGF target genes at 4 hr are repressed by MEK/ERK inhibition, while 28% of PDGF responsive genes are repressed by PI3K inhibition. Interestingly, 12% of genes are repressed by MEK/ERK inhibition and ‘superinduced’ by PI3K inhibition, indicating crosstalk between these pathways. Furthermore, 43% of PDGF responsive genes and 34% of FGF responsive genes are not significantly affected by either inhibitor, which suggests either combinatorial requirement of MEK and PI3K or alternate intracellular pathways drive expression of these genes.
RTK target genes show distinct patterns of effector dependence in a threshold-independent manner.
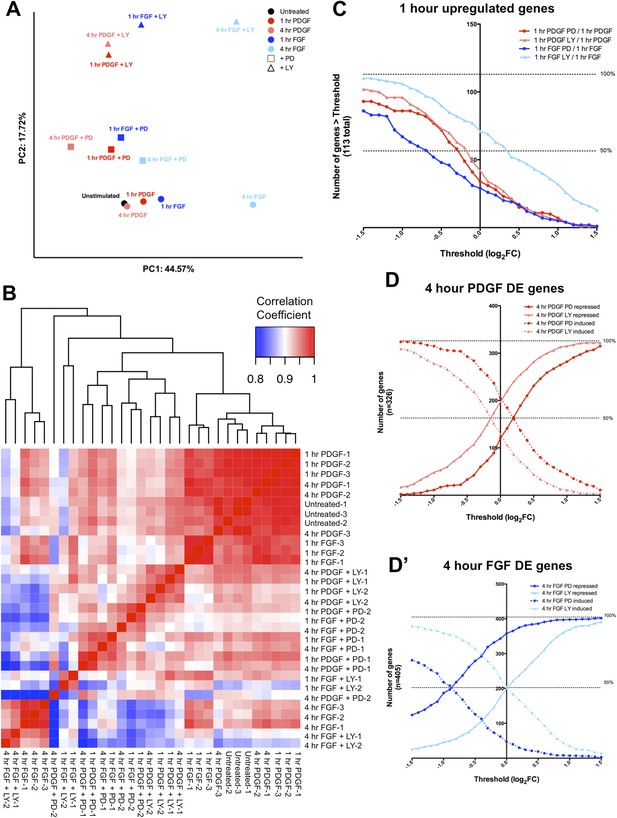
(A) PCA of all 13 conditions highlights effects of ligand stimulation and inhibitor treatment, with the first principal component (44.57% of variance) segregating conditions based on ligand treatment and the second principal component (17.72% of variance) separating samples based on inhibitor treatment. (B) Correlation matrix comparing the effect of inhibitor treatment across all PDGF and FGF stimulated replicates indicates distinct effects for PD and LY treatment. (C) The FGF-ERK and PDGF-PI3K relationships occur independently of FC threshold selection, indicating there is not a specific effect for ERK or PI3K inhibition based on magnitude. The number of genes repressed in the FGF-PD condition is greater than the number of the genes in the FGF-LY condition for all thresholds, while PDGF shows greater PI3K dependence than FGF, both MEK/ERK and PI3K signaling are utilized by PDGF across all thresholds. A subset of genes show significant ‘superinduction’ in the presence of inhibitor treatment, particularly for PI3K inhibition downstream of FGF. Each point on the x-axis represents a different threshold, and the value on the y-axis indicates the number of genes with a log2 ratio greater than that threshold. (D) The FGF-ERK and PDGF-PI3K relationships at 4 hr are also conserved independently of FC threshold selection. For any given threshold, (D) PI3K inhibition represses more PDGF target genes and MEK/ERK inhibition induces more PDGF target genes. Similarly, (D') MEK/ERK inhibition has a stronger repressive effect on FGF target genes than PI3K inhibition at all thresholds.
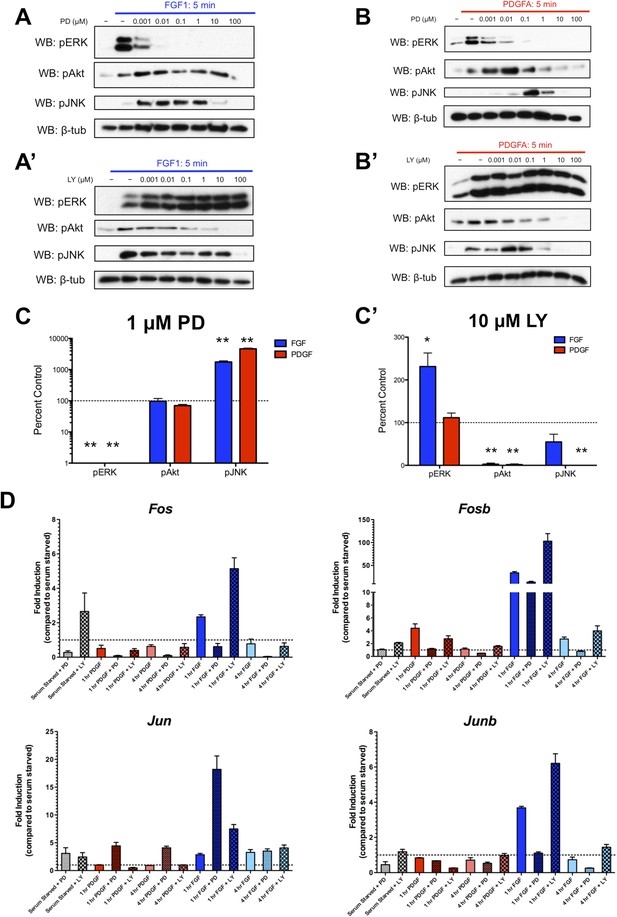
Inhibition of effector activation results in compensatory induction of alternate signaling pathways detectable in the transcriptional response.
(A) PD325901 dose response Western blots reveal induction of pJNK as pERK is progressively inhibited downstream of FGF signaling. (A') Similarly, LY294002 dose response Western blots reveal increased pERK signal as pAkt is inhibited. (B) Inhibitor dose response Western blots in response to PDGF signaling show activation of c-Jun N-terminal kinase (JNK) in response to MEK/ERK inhibition but (B') no activation of ERK following PI3K inhibition. (C) Quantification of effector activation in response to FGF (blue) or PDGF (red) at the doses used in the RNA-seq experiment reflects (C) increased pJNK activation when MEK/ERK signaling is inhibited and (C') increased pERK induction when PI3K activity is blocked. Data plotted as mean ± SEM, n = 3 and compared using two sample, unpaired t-test to baseline of 100% (no change). *p < 0.05; **p < 0.001. (D) Gene expression reflects the crosstalk observed at the signaling level, as verified by qPCR for selected target genes. Canonical ERK targets such as Fos, Fosb, and Junb are ‘superinduced’ upon LY treatment, while the JNK target Jun is ‘superinduced’ with PD treatment. Interestingly, a degree of ‘superinduction’ is observed in the presence of inhibitor prior to growth factor addition, which may reflect RTK-independent crosstalk between intracellular pathways. Data plotted as mean ± SEM, n = 3.
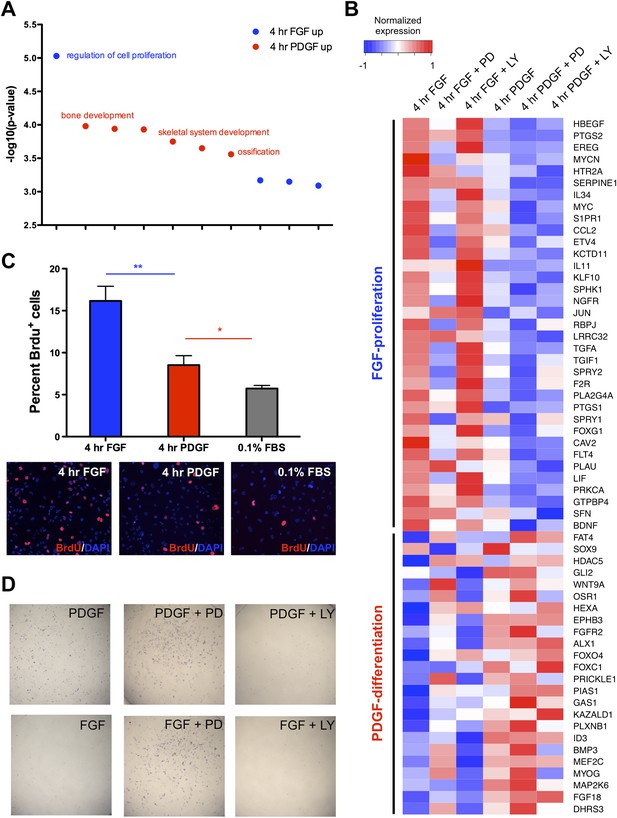
Distinct cellular outcomes are specified in response to PDGF and FGF signaling.
(A) Gene ontology analysis of genes DE between the 4-hr FGF and 4-hr PDGF conditions shows enrichment for regulators of cell proliferation in genes upregulated by FGF treatment. In contrast, genes implicated in skeletal differentiation are overrepresented in the genes more highly expressed following PDGF treatment (B) Genes associated with cell proliferation are strongly upregulated by FGF treatment, while genes associated with cell differentiation are increased following PDGF stimulation. In addition, MEK inhibition represses many proliferation genes but induces differentiation genes, while PI3K inhibition has the opposite effect. Genes ordered by decreasing ratio of FGF:PDGF expression. (C) FGF induces a significantly more robust proliferation response than PDGF in MEPMs, although PDGF does promote a modest response compared to starved cells (0.1% FBS). Quantification plotted as mean ± SEM, n = 3. Two tailed, unpaired t-test: *p < 0.05; **p < 0.005 (D) PDGF, but not FGF, treatment promotes alkaline phosphatase (AP) (osteoblast marker) positive cells. Furthermore, PD treatment drives AP staining, while LY treatment abolishes AP staining independent of growth factor stimulation. AP staining performed 8 hr following ligand treatment.
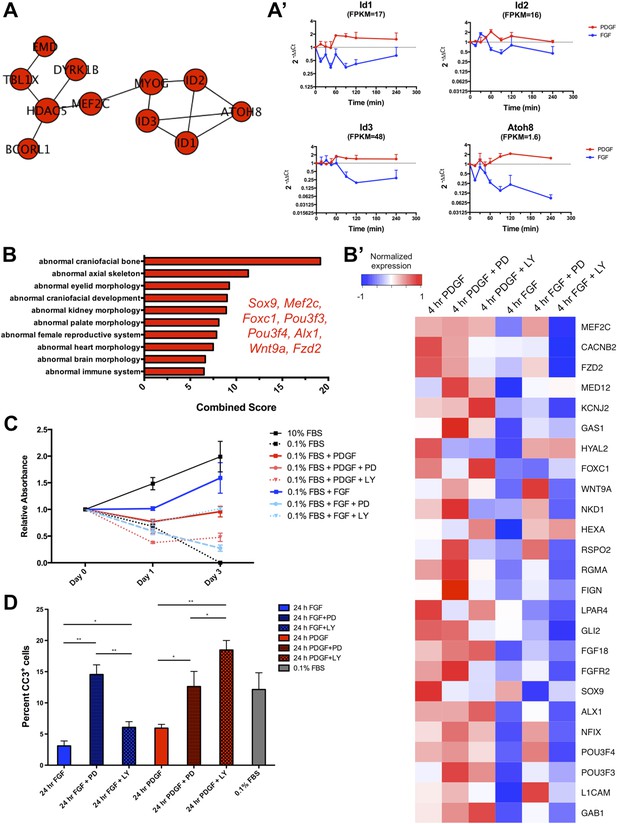
PDGF-mediated differentiation responses exhibit a preference for PI3K signaling, while FGF-mediated effects on proliferation show greater MEK/ERK dependence.
(A). The PPI network for the genes upregulated at 4 hr following PDGF treatment shows a concentration of genes involved in controlling cell differentiation (Id1, Id2, Id3, Atoh8, Mef2c), suggesting a PDGF-mediated differentiation program in MEPMs. (A') Validation of genes in the PDGF target PPI network by qPCR confirmed these genes are increased following PDGF stimulation compared to FGF treatment. (B) Genes with higher expression at 4 hr following PDGF treatment show enrichment for craniofacial and skeletal phenotypes in mice, consistent with the enrichment for differentiation genes in the 4-hr PDGF condition. The top 10 terms are shown, and selected genes with craniofacial phenotypes are listed. (B') Heatmap displaying expression of genes associated with craniofacial phenotypes and exhibiting increased expression at 4 hr PDGF stimulation. Many of these genes show increased expression upon MEK inhibition and decreased expression upon PI3K inhibition, consistent with a PDGF-PI3K-differentiation circuit. (C) Crystal violet assay demonstrates that a full FGF response (blue lines) requires both MEK/ERK and PI3K activity, although the effect of MEK/ERK inhibition is significantly greater. In contrast, the PDGF-mediated effect (red lines) on cell viability requires both MEK/ERK and PI3K activity, suggesting these pathways are utilized somewhat equally downstream of PDGF signaling. Data plotted as mean ± SEM, n = 3. (D) Cleaved Caspase-3 staining reveals that inhibition of FGF-mediated MEK/ERK signaling leads to significantly greater apoptosis, while both MEK and PI3K inhibition downstream of PDGF signaling leads to cell death. Data plotted as mean ± SEM, with cell percentages quantified across three fields of view for three biological replicates. CC3: Cleaved Caspase-3. *p < 0.1; **p < 0.01.
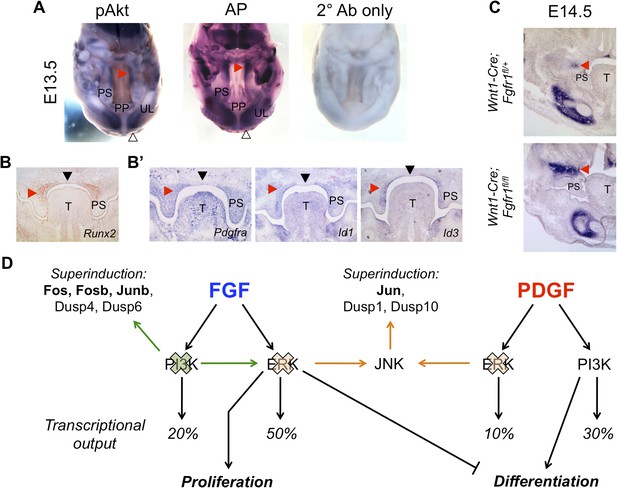
In vivo correlation and perturbation of the RTK-mediated differentiation program during mouse craniofacial development.
(A) At E13.5, pAkt and AP domains co-localize in the middle to posterior secondary palate (red arrowhead) as well as in the developing upper lip (open arrowhead). (B) Domain of Runx2 (osteoblast marker) expression overlaps with (B’) Pdgfra, Id1, and Id3 expression in the middle to posterior palate (red arrowhead), with expression excluded along the midline (black arrowhead). (C) Frontal sections from neural crest conditional Fgfr1 mutants (Wnt1-Cre; Fgfr1fl/fl) exhibit increased AP staining in the developing midface at E14.5, supporting an in vivo role for FGF-mediated repression of osteoblast differentiation (red arrowhead) (n = 3). (D) FGF and PDGF signaling use different signaling pathways to instruct divergent cellular outcomes. FGF drives cell proliferation and represses cell differentiation in an ERK-dependent manner, consistent with a greater percentage of the FGF target genes being MEK/ERK dependent (50%) than PI3K dependent (20%). In contrast, PDGF promotes cell differentiation in a PI3K-dependent manner, and PDGF target genes show greater PI3K dependence (30%) than MEK/ERK dependence (10%). Furthermore, inhibition of PI3K signaling leads to an FGF specific induction of pERK (green) and consequently increased transcription of ERK targets such as Fos, Fosb, and Junb. On the other hand, MEK/ERK inhibition leads to pJNK induction (orange) and transcription of Jun, indicating multiple crosstalk mechanisms across different intracellular pathways in response to RTK activation. PP: primary palate; PS: palatal shelf; T: tongue; UL: upper lip.
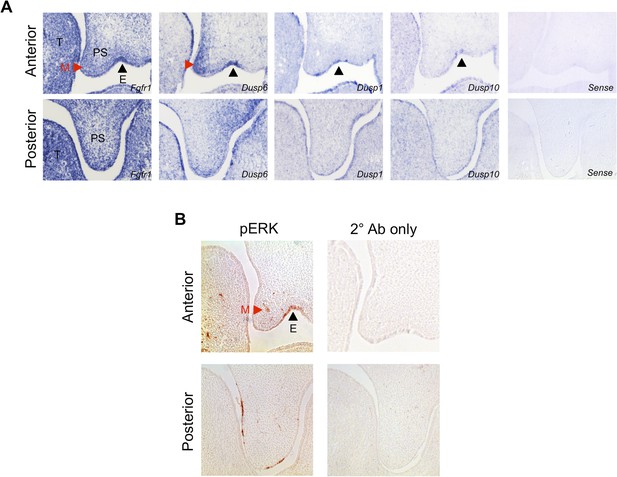
Patterns of gene expression and pERK activity in the E13.5 palate.
(A) In situ hybridization for Fgfr1, Dusp6, Dusp1, and Dusp10 in the E13.5 palate indicates no obvious spatial correlation between receptor expression and putative domains of ERK and JNK activity. Fgfr1 and Dusp6 show enrichment in the anterior palatal mesenchyme (red arrowhead) and epithelium (black arrowhead), while Dusp1 and Dusp10 expression are primarily epithelial (black arrowhead). (B) Immunohistochemistry for pERK reveals broad expression in both the E13.5 anterior palatal mesenchyme (red arrowhead) and epithelium (black arrowhead). E: epithelium; M: mesenchyme; PS: palatal shelf; T: tongue.
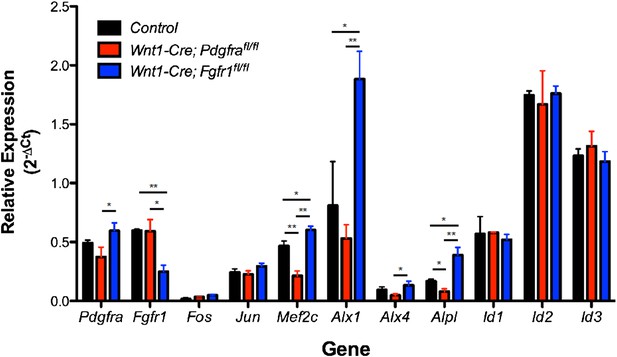
Expression of PDGF and FGF target genes in Pdgfra (red) and Fgfr1 (blue) mutant E13.5 secondary palates compared to control embryos without the Wnt1-Cre transgene (black). *p < 0.1; **p < 0.01, n = 4 for each genotype.
Additional files
-
Supplementary file 1
FPKM values for all genes across all sequenced conditions.
- https://doi.org/10.7554/eLife.07186.014
-
Supplementary file 2
Expression for all differentially expressed genes (Cuffdiff q < 0.1).
- https://doi.org/10.7554/eLife.07186.015
-
Supplementary file 3
Inhibitor dependence for RTK target genes.
- https://doi.org/10.7554/eLife.07186.016
-
Supplementary file 4
Compiled gene ontology analysis.
- https://doi.org/10.7554/eLife.07186.017
-
Supplementary file 5
PCR primers and in situ hybridization probes.
- https://doi.org/10.7554/eLife.07186.018





