A conserved histidine modulates HSPB5 structure to trigger chaperone activity in response to stress-related acidosis
Figures
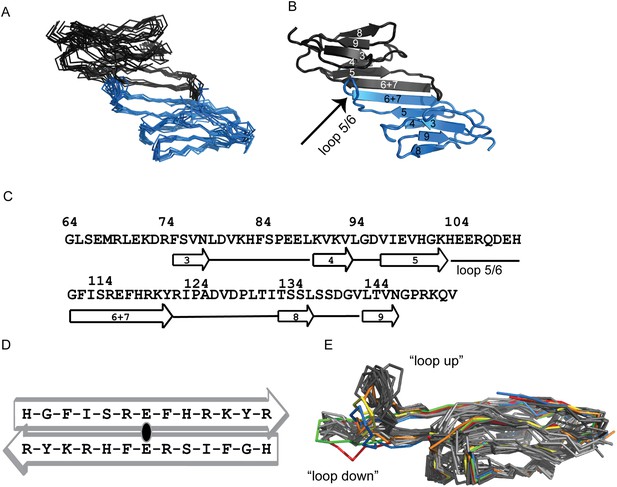
Solution structure ensemble of HSPB5-ACD at pH 7.5 is an anti-parallel dimer.
(A) Backbone traces of ten ACD structures determined by RosettaOligomer are aligned over all residues (RMSD of 1.4 Å). Subunits of the dimer are shown in blue and gray. (B) Cartoon representation of one member of the HSPB5-ACD ensemble is shown. The six strands of the β-sandwich structure are labeled using previously defined nomenclature for ACD structures. The dimer is formed by antiparallel arrangement of the β6+7 strands at the interface. Loop 5/6 is highlighted in the blue subunit. (C) The sequence of the ACD construct for which the solution structure was solved is shown with elements of secondary structure highlighted. (D) The alignment of the β6+7 strands at the dimer interface, shown schematically, places Glu-117 across from Glu-117′. This alignment is called ‘APII’ in previous reports (Clark et al., 2011). (E) Overlay of protomers from all available HSPB5-ACD structures. All members of the solution NMR structure (this paper; PDB 2N0K) and the solid-state NMR structure (2KLR; Jehle et al., 2010) are shown in dark gray and light gray, respectively. Five crystal structures are shown: (1) 2WJ7 (yellow), (2) 3L1G (orange), (3) 4M5S (green), (4) 4M5T (red), and (5) 2Y22 (blue).
-
Figure 1—source data 1
Comparison of the NMR solution structure and published structures.
Backbone root mean square deviations (rmsd) between the solution structure and published structures and the correlation between experimentally measured residual dipolar coupling (RDC) values and those calculated for each published structure (PALES Correlation Coefficient) are given. Values in parentheses are calculated without including loop regions between the β-strands. Protomer–protomer and dimer–dimer rmsd between 2KLR and solution structure ensembles are the averages between all pairwise combinations of all structures in each ensemble. Rmsd values between the solution structure ensemble and the crystal structures are the average of all pairwise rmsd between the ten structures in the ensemble and the crystal structure. 2KLR: solid-state NMR structure; 2WJ7:crystal structure at pH 9.0; 3L1G:crystal structure at pH 4.6; 4M5S: crystal structure at pH 6.0.
- https://doi.org/10.7554/eLife.07304.004
-
Figure 1—source data 2
Multiple sequence alignment of the ten human sHSPs.
ClustalOmega was used to align the sequences. Structural features of HSPB5-ACD are highlighted as follows: ACD is denoted by the blue line; His-104 is identified by the blue arrow; Loop 5/6 is shown in red box; dimer interface is shown in green box. All ACD histidine residues are shown in bold font.
- https://doi.org/10.7554/eLife.07304.005
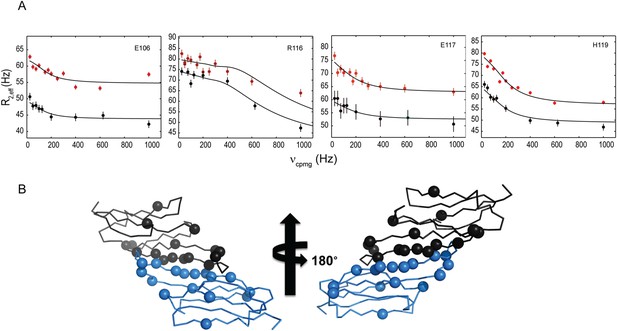
15N-CPMG relaxation dispersion experiments detect dimer-to-monomer exchange.
(A) Relaxation dispersion measurements reveal a two-state transition. Representative relaxation dispersion curves of 15N transverse relaxation rate (R2,eff) plotted as a function of field strength, (νCPMG) are shown (see Materials and methods for details). Data were recorded at static field strengths of 800 MHz (red) and 600 MHz (black) at pH 7.5 and 22°C. Values of kex, δω, and pb were extracted using the program, GUARDD. (B) Backbone representation of HSPB5-ACD dimer is shown with the Cα atoms of exchanging residues that are well fit by a two-state model shown as spheres. Resonances showing relaxation rates in the range 760 s−1 to 1119 s−1 occur mainly at the dimer interface and in Loop 5/6.
-
Figure 2—source data 1
Parameters from relaxation dispersion experiments performed on 0.7 mM HSPB5-ACD at 22°C and pH 7.5.
- https://doi.org/10.7554/eLife.07304.009
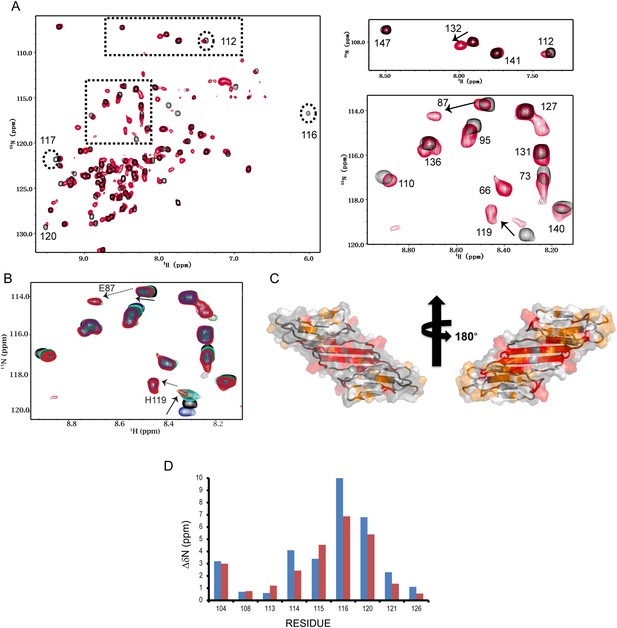
HSPB5-ACD undergoes a conformational transition between pH 7.5 and 6.5.
(A) 1H-15N HSQC spectra acquired on a 200 µM sample of HSPB5-ACD at pH 7.5 and 6.5 (22°C) reveal two states. Full spectra collected at pH 7.5 (black) and 6.5 (red) are overlaid (left). At pH 7.5, 82/85 non-proline residues are observed (residues G64, L65, and S139 are not detected due to fast exchange with H2O). At pH 6.5, approximately ∼65 additional peaks appear, indicating the presence of two conformations. Boxed regions shown on the right provide clear examples of peak doubling. Some resonances (labeled in the panel on the left) disappear from their original positions at pH 7.5 and their new positions could not be determined by inspection of the spectrum at pH 6.5. (B) Example of a full pH titration series (spectra collected at pH 8.4 [blue], 7.5 [black], 7.0 [cyan], 6.7 [green], and 6.5 [red]). The behavior of the resonance of H119 as a function of pH illustrates two pH-dependent processes (see text). Its chemical shift is in fast exchange from pH 8.4 to 7.5 (solid arrow) and changes direction and is in slow exchange from pH 7.0 to 6.5 (dashed arrow). The region shown contains several other resonances that undergo slow-exchange transitions over the same pH range. (C) Residues that undergo the slow exchange transition are highlighted in color on a surface representation of the ACD dimer. Residues with perturbations > 0.2 ppm are red; those with perturbations between 0.1 - 0.2 ppm are orange. (D) Analysis of relaxation dispersion data yields the difference in 15N chemical shift between the major and minor species (ΔδNcalc). The values of ΔδNcalc (blue) are compared to the experimental values obtained from the difference of 15N chemical shifts at pH 7.5 and 6.5, i.e., ΔδN(7.5−6.5) (red), in the histogram. Concordance between these two parameters supports the notion that the minor form detected by relaxation dispersion experiments is the monomeric form of ACD that is populated at lower pH.
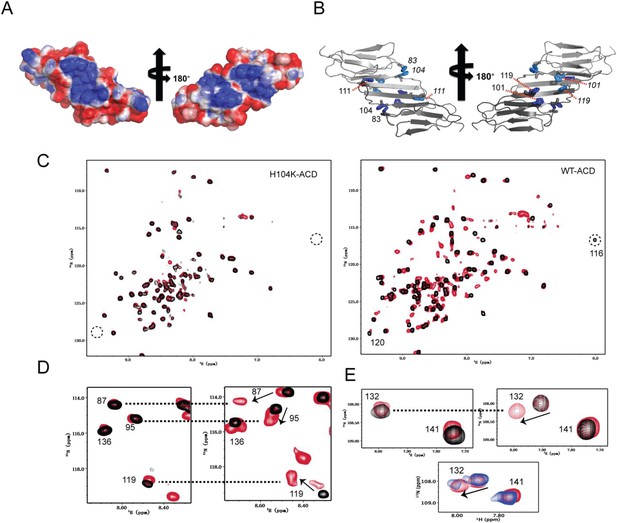
His-104 plays a key role in the dimer-monomer transition.
(A) The electrostatic surface of HSPB5-ACD at pH 7.5 (calculated using experimentally determined histidine pKR values) reveals patches of positive (blue) and negative (red) charges that cross the dimer interface. (B) The five histidines of HSPB5-ACD (blue and cyan sticks) occur as pairs and are located in proximity to the dimer interface. (C) Mutation of His-104 shifts the dimer-monomer equilibrium as observed by NMR. Overlays of 1H-15N HSQC spectra at pH 7.5 (black) and pH 6.5 (red) are shown for H104K-ACD (left panel) and WT-ACD (right panel). A single set of peaks is observed in WT-ACD at pH 7.5, and peaks due to the monomer conformation appear as the pH is lowered. Peaks belonging to the dimer interface, e.g., R120 and R116 disappear from their original positions in WT-ACD at pH 6.5 and in H104K-ACD at both pH conditions (dotted circles). (D, E) Expanded regions of 1H-15N HSQC spectra of WT-, H104K-, and H104Q-ACD. The same color scheme is used for WT- and H104K-ACD as in panel C; H104Q-ACD overlay is in blue (pH 7.5) and red (pH 6.5). (D) Comparison of H104K-ACD (left panel) and WT-ACD (right panel). Dotted lines connect resonances in similar positions in H104K-ACD and WT-ACD. The example shows that the peaks for residues 87, 95, 119, and 136 have similar positions in H104K-ACD (at both pH values) as in WT-ACD at pH 6.5. (E) Comparison of identical regions of spectra of H104K-ACD (top left panel), WT-ACD (top right panel), and H104Q-ACD (lower panel). Both forms (dimer and monomer) are observed in the H104Q-ACD spectrum at both pH values, whereas only peaks corresponding to the monomer are observed in H104K-ACD at both pH values.
-
Figure 4—source data 1
The pKR values and tautomeric states of HSPB5-ACD histidine side-chain imidazole rings (22°C) are listed.
n.d.: not determined because resonances broaden below pH 7.5 and become undetectable.
- https://doi.org/10.7554/eLife.07304.013
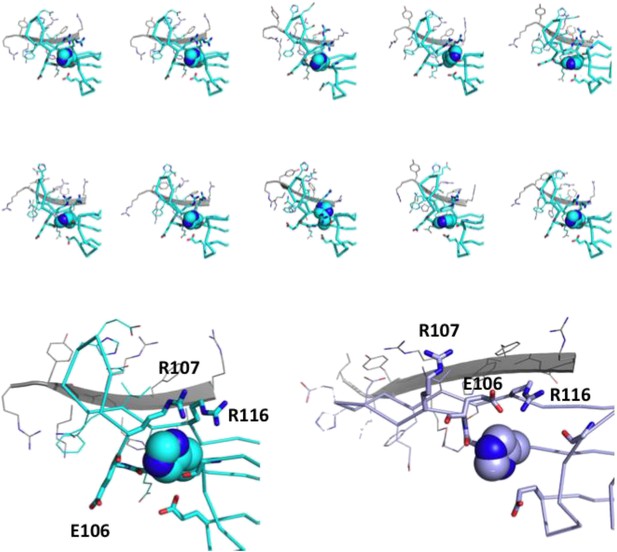
His-104 is at the center of a dynamic network of charged and H-bonding interactions.
Top panel: ten individual members of the solution ensemble are shown. One protomer of the dimer is in cyan; the other protomer is in gray. His-104 is shown as space-filling spheres; carbonyl groups that are in proximity to His-104 are shown in stick representation; side chains that are proximal to His-104 are shown in stick representation; side chains in Loop 5/6 and in the dimer interface as shown as lines; Bottom panel: examples of a loop-up (shown in cyan; this publication PDB 2N0K) and loop-down conformation (shown in violet; from 2KLR) are shown to illustrate the rearrangement of potential interacting partners in the two conformations. As above, the second subunit in a dimer is shown in gray.
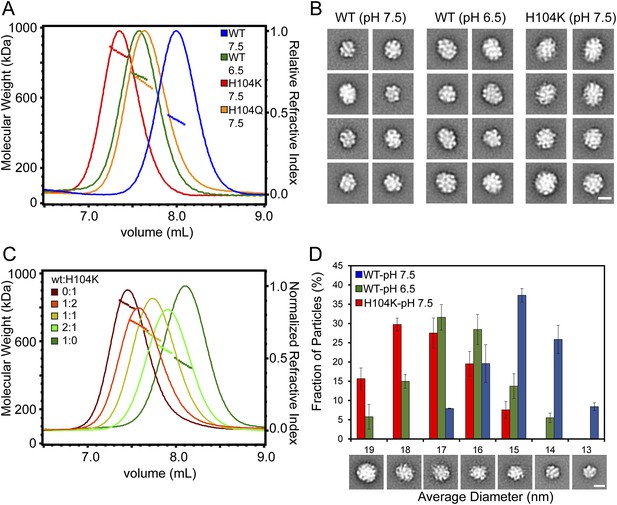
Destabilizing the ACD dimer interface via low pH or His-104 mutation triggers large expansion of HSPB5 oligomers.
(A) SEC-MALS analysis showing protein elution profile (refractive index, right Y-axis) with average Mw (horizontal trace under peak corresponds to left Y-axis) for WT-HSPB5 at pH 7.5 (blue) and pH 6.5 (green), and H104Q-HSPB5 (orange) and H104K-HSPB5 (red) at pH 7.5. (B) 2D projection class averages showing representative ensemble of oligomer sizes. (C) SEC-MALS analysis showing elution profile and average Mw of mixed oligomers of WT and H104K-HSPB5 incubated together at ratios of 1:0 (green), 2:1 (light green), 1:1 (yellow), 1:2 (orange), and 0:1 (dark red), respectively. (D) Histogram of oligomer diameters showing fraction of oligomer particles vs average diameter (nm) for WT at pH 7.5 (blue) and pH 6.5 (green), and H104K at pH 7.5 determined by negative-stain EM 2D classification. Representative 2D class averages corresponding to measured diameter are shown. Experiments were performed in triplicate. Scale bar (lower left image) equals 10 nm.
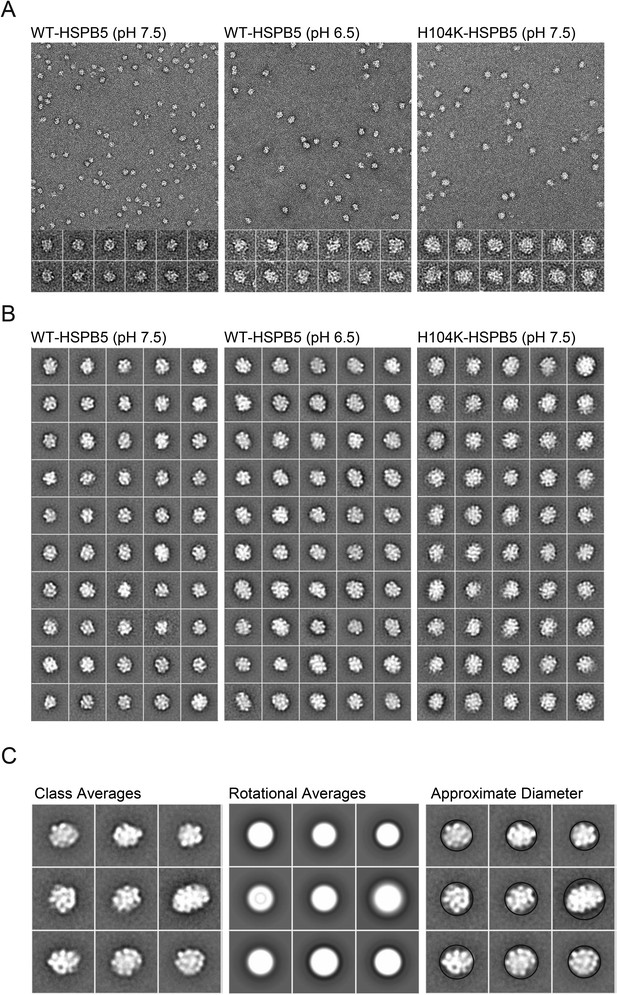
EM micrograph images, 2D classification, and particle diameter estimation for WT and H104K HSPB5.
(A) Representative micrograph images of WT-HSPB5 at pH 7.5 and 6.5 and H104K-HSPB5 at pH 7.5 negatively stained with 0.75% uranyl formate (scale bar equals 20 nm). Example individual particle projections are shown below with scale bar equal to 10 nm. (B) 2D reference free class averages for WT-HSPB5 pH 7.5, WT-HSPB5 pH 6.5 and H104K-HSPB5 pH 7.5 from data sets of 5,321, 5713, and 5145 single particles for respectively. Scale bar equals 10 nm. (C) Example class averages, corresponding rotational averages and estimated diameter (nm) for HSPB5 oligomers.
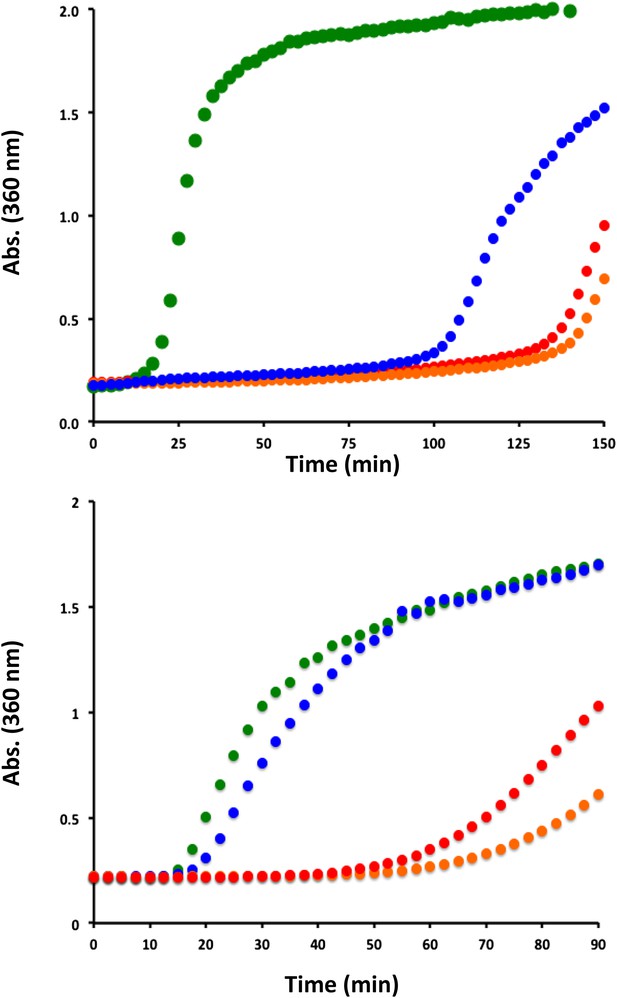
His-104 mutants of HSPB5 are effective at delaying the onset of aggregation of a model client protein.
(Top panel) Aggregation of DTT-denatured bovine αLactalbumin at 42°C in the absence (green) and presence of WT-HSPB5 (blue) or HSPB5 mutants H104K (red) and H104Q (orange). Light scattering at 360 nm was used to monitor the DTT-induced aggregation of αLac (600 µM) in the presence and absence of HSPB5 (40 µM). Assays were performed in duplicate and the average scattering curves are shown. (Bottom panel) Aggregation of Yeast Alcohol Dehydrogenase in the presence of EDTA and DTT, at 37°C in the absence (green) and presence of WT-HSPB5 (blue) or HSPB5 mutants H104K (red) and H104Q (orange). Light scattering with 360 nm light was used to monitor ADH (100 µM) aggregation in the presence and absence of HSPB5 (20 µM). Assays were performed in duplicate and the average scattering curves are shown.
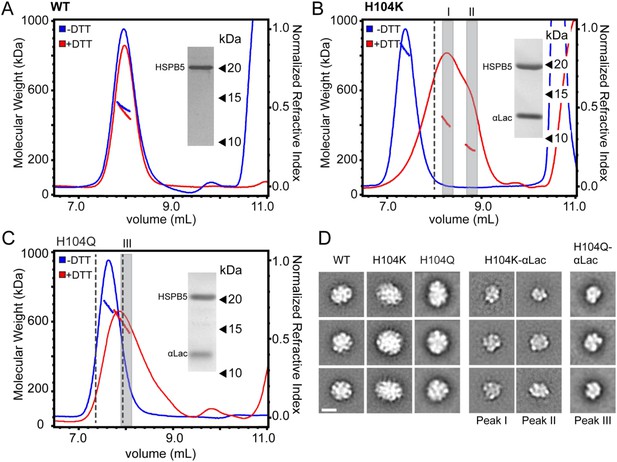
H104K- and H104Q-HSPB5 oligomers reorganize into small, long-lived client-bound complexes in the presence of αLac model client protein.
(A–C) SEC-MALS analysis and corresponding Mw of WT (A), H104K (B) and H104Q (C), HSPB5 oligomers (40 μM in subunit concentration) incubated with αLac (120 μM) in the absence (blue) and presence (red) of 50 mM DTT to destabilize αLac. SDS-PAGE analysis of peak fractions for corresponding HSPB5-αLac incubations with DTT (red trace) is shown (inset). Dashed lines correspond to WT elution at 8 ml and H104K elution at 7.4 ml. (D) 2D projection class averages of H104K- and H104Q-HSPB5-αLac peak fractions corresponding to peaks I, II, and III (gray bars) in B and C are shown with representative averages of WT, H104K-, and H104Q-HSPB5 incubated without αLac substrate for comparison. Scale bar (lower left panel) equals 10 nm.
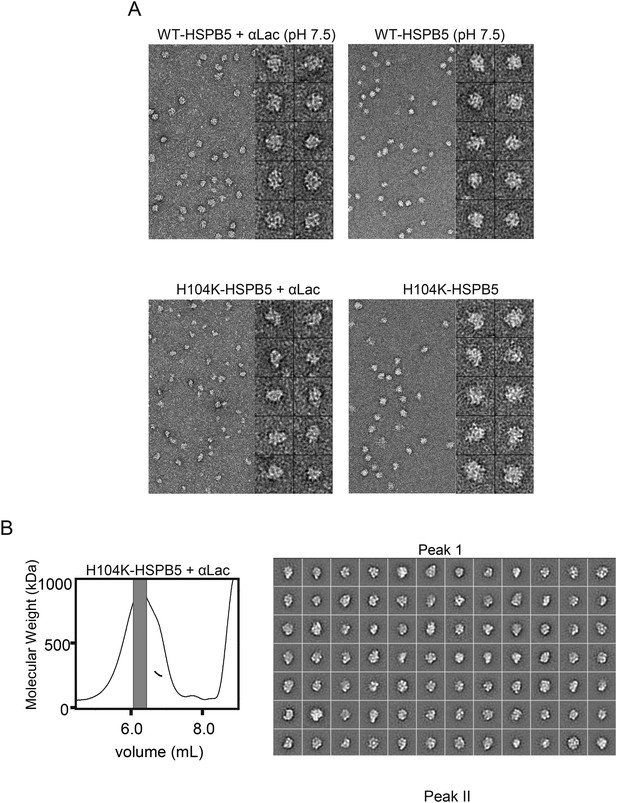
EM micrograph images and 2D classification of HSPB5 incubated with αLac.
(A) Representative micrograph images and single particles of WT-HSPB5 and H104K-HSPB5 incubated with or without αLac substrate and DTT prior to fractionation by SEC-MALS showing the shift in oligomer size only occurs with H104K-HSPB5 and α-Lac. Scale bars equal 20 nm and 10 nm for the micrographs and single particles, respectively. (B) Complete set of 2D projection class averages of H104K-HSPB5-αLac isolated in Peak 1 and Peak II following SEC-MALS fractionation. Class averages were obtained from 11,727 particles in fraction I and 9257 particles in fraction II are shown and the scale bars equal 10 nm.
Tables
NMR data and refinement statistics for HSPB5-ACD structures
| NMR distance and dihedral constraints | HSPB5-ACD |
|---|---|
| Distance constraints | |
| Total NOE | 838 |
| Intraresidue | 310 |
| Inter-residue | |
| Sequential (|i − j| = 1) | 255 |
| Medium-range (|i − j| < 4) | 85 |
| Long-range (|i − j| > 5) | 188 |
| Inter-molecular | 36 |
| Total dihedral angle restraints | |
| φ (TALOS) | 72 |
| ψ (TALOS) | 72 |
| Residual Dipolar Couplings (RDCs) | |
| 1H-15N RDCs | 81 |
| Structure statistics | |
| Violations (mean ± s.d.) | |
| Distance constraints (Å) | 0.48 ± 0.45 |
| Dihedral angle constraints (°) | 14.4 ± 14.7 |
| Average pairwise r.m.s deviation (Å)* | |
| Heavy | 2.46 ± 0.97 |
| Backbone | 1.48 ± 0.6 |
-
*
Average pairwise r.m.s.d. was calculated among ten refined structures.
Dimer-to-monomer dissociation constants for HSPB5-ACD determined by ITC*
| Temperature | pH | Kd (mM) | ΔH (cal/mol) |
|---|---|---|---|
| 25°C | 7.5 | 0.002 ± 0.002 | 8772 ± 3960 |
| 25°C | 6.5 | 0.030 ± 0.016 | 3242 ± 432 |
| 37°C | 7.5 | 0.036 ± 0.002 | 9328 ± 527 |
-
*
See ‘Methods and materials’ for experimental details and data analysis.





