Spatially coordinated dynamic gene transcription in living pituitary tissue
Figures
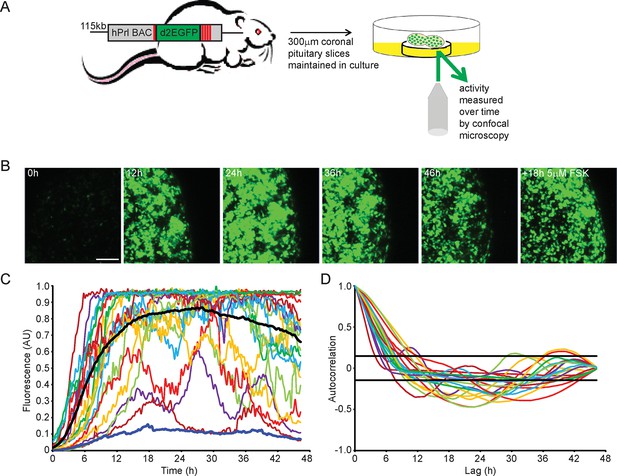
Patterns of prolactin gene transcription activity in pituitary tissue.
(A) Schematic of the reporter construct and experimental approach used. (B) Images of d2EGFP expression in lactotroph cells of adult male pituitary tissue during a 46-hr imaging period, in basal culture media, and a subsequent 18-hr imaging period following stimulation with forskolin (FSK). Images shown are a maximum intensity projection of a z-stack over 242 µm. Bar represents 100 µm. (C) Graph of d2EGFP fluorescence (average intensity, arbitrary units) from 20 individual cells (representative of 101 cells analysed, from one experiment) from adult male pituitary tissue. The black line represents the mean activity from all the cells analysed. The dark blue line represents the background fluorescence intensity (mean from five areas). (D) Autocorrelation analysis of d2EGFP fluorescence intensity from cells shown in (C), with the 95% confidence interval representing a white noise process shown between black lines. Data are representative of three independent experiments. For validation of single cell imaging see Figure 1—figure supplement 1. d2EGFP, destabilised EGFP.
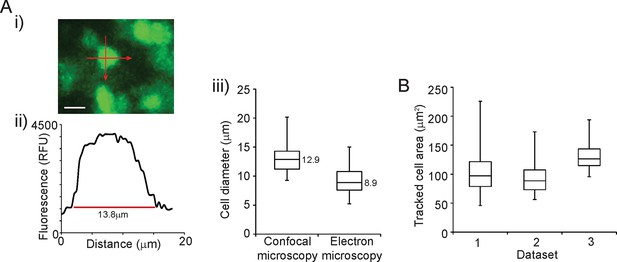
Validation of single cell imaging.
(A) Cell diameters were measured in two planes by confocal microscopy, as shown (i-ii) and compared to estimates of cell diameters from electron microscopy (iii). Estimates of cell diameter were slightly smaller by electron microscopy, which may be accounted for by the processing of tissues for electron microscopy. Estimates of cell diameter are within the range of a typical eukaryotic cell, indicating that settings used for fluorescence microscopy gave good image resolution. (B) Areas of cells tracked from live-cell imaging are very similar to estimated cell areas calculated from cell diameters in (A,iii) (99–160 μm2 quartile range). The similar distribution of cell areas across three independent data sets shows the consistency of our cell tracking approach. Box plots show the median, interquartile range, min, and max values. Bar in image represents 10 µm.
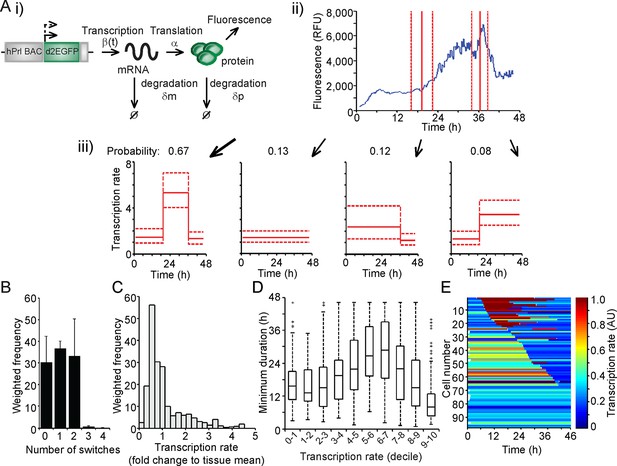
Characterisation of prolactin transcription dynamics.
(A) i) Schematic of the parameters (translation rate, mRNA and protein degradation rates) used by the stochastic switch model to back-calculate to the original transcription rate, β(t), of the reporter construct. For each fluorescence time series, there exist several mutually exclusive possible transcriptional profiles based on the sampled switch times. ii) The graph shows the relationship between the measured fluorescence activity (blue) and possible switch times (red, dashed lines represent ± 1.96 SD). iii) The four possible profiles associated with the estimation of two transcriptional switches are shown along with their predicted probability of occurrence. (B–E) Characterisation of transcription rate activity of the reporter construct in cells maintained within adult pituitary tissue. (B) The number of switches in transcriptional states estimated during the imaging time-course. The frequency of switches is weighted by the probability of occurrence of each profile. Data are represented as the mean + SD from three independent experiments. (C) Distribution of the frequency of transcription rates estimated from fluorescence activity, with a weighted density calculated from all possible transcriptional profiles for each cell. The cumulative distribution of transcription rates from three independent experiments is shown in Figure 7B. (D) The duration of transcriptional states, with the caveat that the duration is the minimum duration as complete periods of activity were not detectable for most states. Transcription rates were binned into deciles and the associated minimum durations calculated. Boxplots are calculated by random sampling of the weighted transcription rate distributions for each cell. There is some evidence for longer durations at mid-range transcriptional rates (4th–8th decile), although longer observation windows may yield further trends. Data shown were pooled from three independent experiments. Boxplots represent the median and interquartile range (IQR), with whiskers drawn 1.5xIQR away from the lower and upper quartile. (E) Heatmap of cell transcription rate patterns ordered by the timing of the first switch event. A single transcriptional profile was selected at random for each cell taking into account the probability of occurrence. Data shown in (C) and (E) are from a single experiment on adult pituitary, representative of three independent experiments. SD, standard deviation.
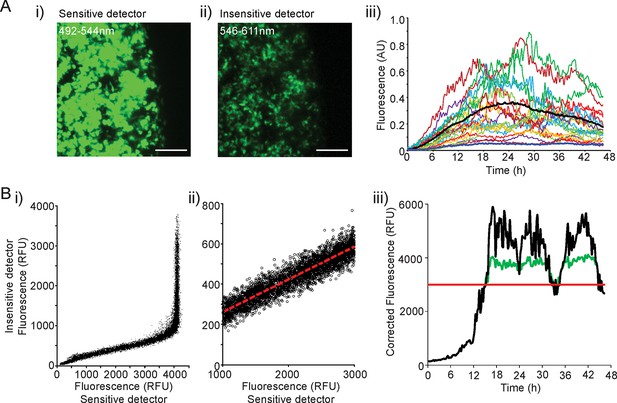
Processing of fluorescence time-series prior to transcription rate estimation.
(A) The range of fluorescence activity resulting from hPRL gene expression exceeded the detection range of the microscope systems used. Data saturation would affect the transcription rates and timing of switches between different transcription rates estimated from the stochastic switch model. To overcome this issue, we collected d2EGFP signal at 492–544 nm and 546–611 nm wavelengths simultaneously using two detectors at different levels of sensitivity. Comparisons of signal intensity are shown in images i) and ii) and graph of expression patterns measured using the insensitive detector in iii), which is comparable to the graph shown in Figure 1C for the sensitive detector. Bars represent 100 µm. (B) i) and ii) Fluorescence intensity measurements from the two detectors were combined using linear regression analysis. iii) The combined data (black line) consists of data from the sensitive detector (green line) until a threshold value (red line), after which data from the less sensitive detector, rescaled using the linear regression equation, was used. hPRL, human prolactin.
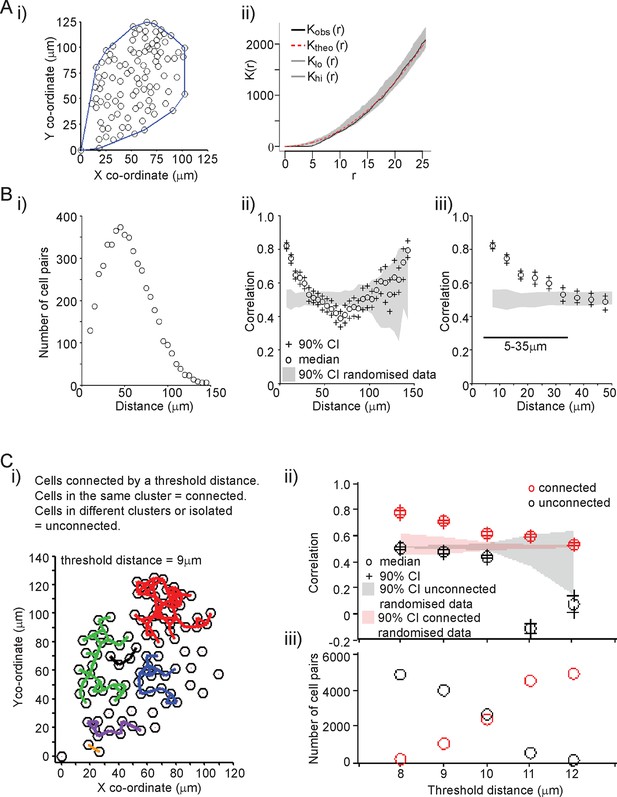
Spatial organisation of transcription activity in pituitary tissue.
(A) Assessment of the spatial distribution of lactotroph cells with PRL gene transcriptional activity. i) The median position of the cells over the time-course along with the field or convex hull occupied by the cells. ii) Ripley’s k function was used to test whether the cellular distribution was random. The observed distribution (Kobs (r)) deviates from the theoretical prediction (Ktheo (r)) in the short r range due to the finite size of the cells, above which the cell distribution is found to be indistinguishable from random and within the 95% confidence interval (Klo (r) – Khi (r)). (B) Correlation vs distance analysis of cells from adult male pituitary tissue. The correlation between two time series xi and yi measured at N times is defined as . i) The distribution of distances between cell pairs. Correlation vs distance plots ii) all cell data and iii) cell pairs within 50 µm; indicate that proximally located cells have more similar activity than cells located further apart. The median and 90% confidence interval (CI) are shown along with the profile obtained from the 90% confidence interval of randomised of cellular transcription patterns. The range shown indicates the distances over which the randomised data were significantly greater (paired t-test, p<0.001) to the non-randomised data. (C) Analysis of correlation mediated by a potential lactotroph cell network in adult male pituitary tissue. i) Cell connectivity map with cells classified as connected or unconnected according to the threshold distance used. Connections between cells at a single time point are portrayed. ii) Correlation was calculated between connected (red) and unconnected cells (black) in parallel with randomised data. iii) The number of connected and unconnected cell pairs at various threshold distances. Connected cells were more correlated in their transcription activity than unconnected cells. Cell connectivity in ii)-iii) are based on median cell distances across the time-course to take into account cell movement. Data shown are representative of three independent experiments. PRL, prolactin.
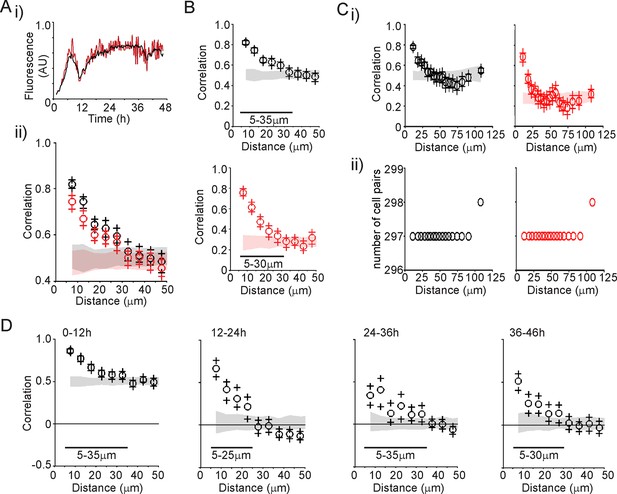
Characterisation of spatial organisation of prolactin transcription activity.
(A) Spatial correlation between proximally positioned cells was not due to restricted resolution in the imaging data. The regions of interest (ROI) analysed were collapsed to 25% of the original area to ensure correlation was not due to overlap of signal between cells. i) An example cell shows that this method caused the fluorescence signal to show large fluctuations in intensity but with the pattern of activity maintained. This is probably due to less averaging of signal intensity due to the smaller number of pixels. ii) The pattern of correlation over distance was maintained for both sizes of ROIs (100% ROI in black and 25% ROI in red) showing that imaging resolution was not a factor in causing increased correlation between proximally located cells. (B) Spatial correlation was maintained when imaging was performed at differing levels of sensitivity (as discussed in Figure 2—figure supplement 1). The correlation vs distance analyses show similar results for the two levels of signal detection (sensitive detector shown in black, insensitive detector shown in red), although the absolute value of correlation changes due to the inclusion of either saturated values or zeros. Ranges shown indicate the distances over which the randomised data were significantly greater (paired t-test, p<0.001) to the non-randomised data. Data were calculated in bins of 5 µm. (C) Spatial organisation of transcription was maintained when the number of cell pairs in each bin was normalised. Correlation vs distance plots are shown (i) along with the number of cell pairs in each bin (ii). Graphs are coloured as in (B). (D) Spatial organisation of transcription activity is maintained over 48 hr. Correlation of transcription patterns was not solely due to the stimulation of cell activity by the removal of dopamine as this correlation was maintained between proximally located cells over 48 hr and after 24 hr when hPRL-d2EGFP reporter gene activity declined. Data shown are from the sensitive detector, although similar results were obtained with data from the insensitive detector (not shown). Correlation vs distance plots are shown as described in Figure 3B.
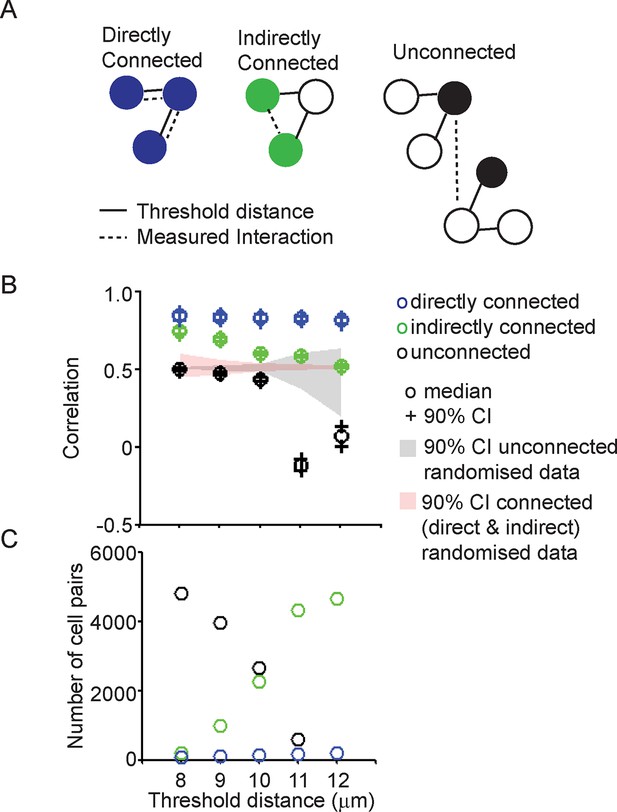
Correlation of transcription profiles within a cellular network structure.
(A) Schematic showing the classification of cells into directly connected, indirectly connected and unconnected cell groups. (B). Analysis of correlation between different cell groups (classified as pictured in A) with connections calculated using a defined threshold distance. Correlation was calculated in parallel with randomised data. (C) The number of directly connected, indirectly connected, and unconnected cell pairs at various threshold distances. Directly connected cells were more correlated in their transcription activity than indirectly connected or unconnected cells. Cell connectivity in (B) and (C) are based on median cell distances across the time-course to take into account cell movement. Data shown are from the same experiment in Figure 3C and are representative of three independent experiments.
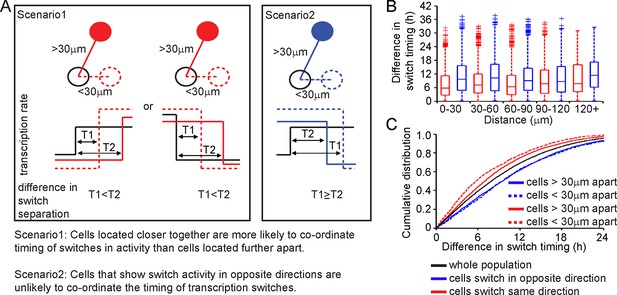
Spatial organisation of stochastic switch model derived prolactin transcription dynamics.
(A) Schematic outlining the hypothesis that was used to assess the spatial organisation of PRL transcription dynamics. The hypothesis was that two cells located closer together will tend to switch transcription in the same direction with more synchronous timing than cells that are located further apart. Moreover, a similar co-ordination in the timing of switches will not be observed if switches occur in the opposite direction. Comparisons are made to the index cell (black). Red denotes cells that switch transcription rate in the same direction, blue denotes cells that switch transcription rate in the opposite direction. T1 is the time interval between cells located within 30 μm (and dashed lines) and T2 is the time interval between cells located more than 30 μm apart (and solid lines). (B) Graph showing boxplots of switch timing intervals in cells that switch in the same direction and cells that switch in different directions, binned by the distance between cells. A rising trend is seen in the time interval between transcription rate switch events in cells that switch activity in the same direction (red), but not in cells that switch activity in the opposite direction (blue). Specifically, the median time interval between switch events is smallest in cells that are located within 30 μm and that switch activity in the same direction. Cumulative distributions and significance testing of these differences are shown in (C). All pairwise switches are considered. Boxplots represent the median and interquartile range (IQR), with whiskers drawn 1.5xIQR away from the lower and upper quartile. (C) The cumulative distribution of the time interval between switch events shows that cells within 30 µm that switch activity in the same direction (red dashed line) do so within a smaller time frame than cells located greater than 30 µm apart (red solid line), the unsorted population (black) and cells that switch activity in opposite directions (blue dashed and blue solid lines) (confirmed by significant p-value <0.01 of Kolmogorov-Smirnov tests). These were calculated by sampling at random, a pair of possible transcriptional profiles for each pair of cells. Data shown were pooled from three independent experiments.
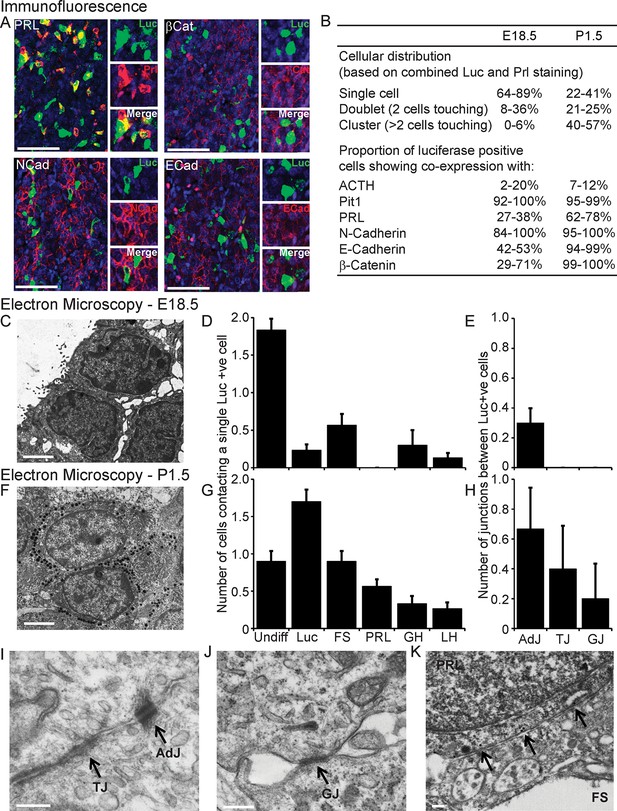
Characterisation of cell signalling potential in immature pituitary glands.
(A, B) Immunofluorescence analysis of lactotroph cell connectivity in developing pituitaries. (A) Immunofluorescence images show co-expression of luciferase antibody (green), used to identify lactotroph cells, with PRL, N-Cadherin, E-Cadherin, and β-catenin (red) in paraffin-embedded sections from P1.5 PRL-Luc49 rats. Nuclei were counterstained with DAPI (blue). Right: 8x crop images. Bars in images represent 50 µm. (B) Table showing the proportion of lactotroph cell clustering in E18.5 and P1.5 pituitary tissue and the level of co-expression of the luciferase protein and the protein of interest indicated, counted from immunofluorescence images. (C–K) Electron microscopy analysis of lactotroph cell connectivity in E18.5 (C-E) and P1.5 (F-K) pituitaries. (C, F) Representative image of two luciferase positive cells detected through immunogold labelling of luciferase antibody. Bars represent 1 µm. (D, G) Graph of the number of cells contacting an individual luciferase-positive cell. Undiff, undifferentiated cell. Luc, luciferase-expressing cell. FS, folliculostellate cell. PRL, lactotroph. GH, somatotroph. LH, gonadotroph. (E, H) Graph quantifying the different types of cell junctions present between luciferase-positive cells. Data in D-E and G-H are represented as mean + SEM. (I, J) Electron micrograph of a visually normal adherens junction (AdJ), a visually normal tight junction (TJ) and a visually normal gap junction (GJ) in P1.5 pituitaries. Bars represent 200 nm. (K) Electron micrograph of an abnormal adherens junction in P1.5 pituitary. Bar represents 200 nm. Information and validation of antibodies used are presented in Figure 5—figure supplement 1. SEM, standard error of the mean.
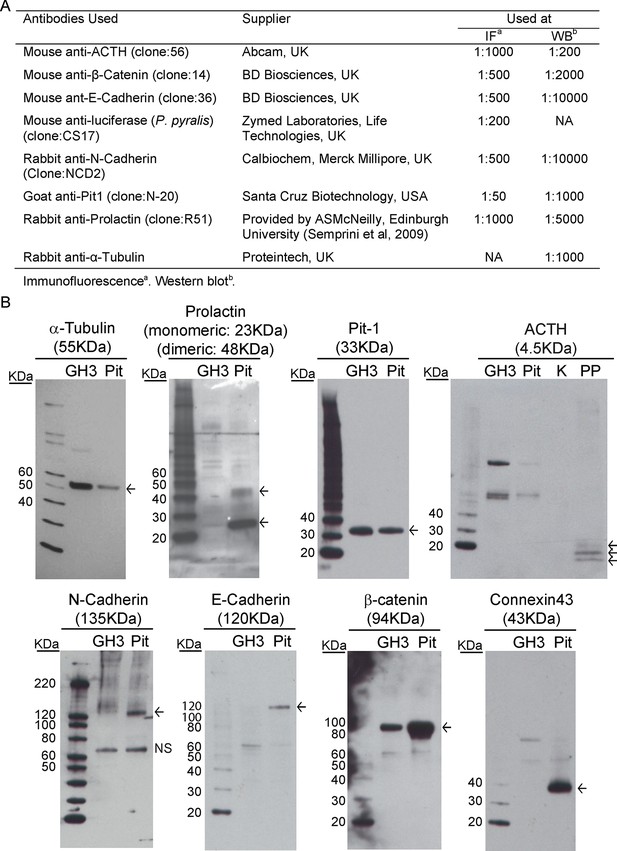
Description and validation of antibodies used.
Information and validation of antibodies used in analyses presented in Figure 5 and Figure 8—figure supplement 1. (A) Table describing antibodies used. (B) Antibodies were validated by western blot to determine the specificity of the antibody and to ensure a protein of the appropriate size was detected. NS = non-specific binding.
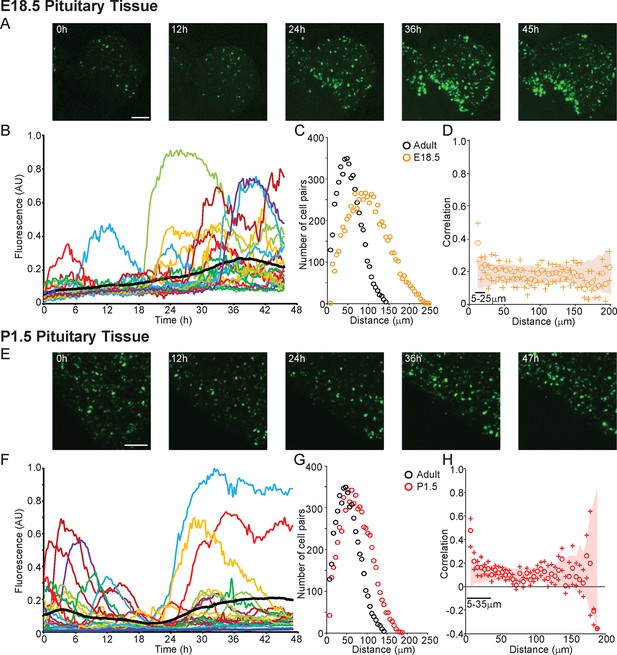
Patterns and spatial organisation of prolactin gene transcription activity in immature pituitary tissue.
(A, B) Activity of the hPRL-d2EGFP reporter construct in single cells in E18.5 pituitary tissue over 46 hr. (A) Images of d2EGFP expression in lactotroph cells in E18.5-day-old pituitary tissue (male). (B) Fluorescence profiles from 20 individual cells, representative of 136 cells analysed (average intensity, arbitrary units). The black line represents the mean average activity from all the cells analysed (136 cells), the dark blue line represents the background fluorescence intensity (mean from five areas). (C, D) Spatial correlation between fluorescence profiles of PRL transcription activity is reduced in E18.5 pituitary tissue in comparison to adult tissue. (C) The distribution of cell pairs in E18.5 tissue is compared to the adult cellular distribution. (D) Correlation vs distance plots show no large differences between randomised and non-randomised data over short cell-to-cell distances. (E, F) Activity of the hPRL-d2EGFP reporter construct in single cells in P1.5 pituitary tissue over 46 hr. (E) Images of d2EGFP expression in lactotroph cells in P1.5-day-old pituitary tissue (female). (F) Fluorescence profiles from 20 individual cells, representative of 115 cells analysed. Lines are coloured as in (A) with the mean representing the activity from all cells analysed. (G, H) Spatial correlation between fluorescence profiles of PRL transcription activity was reduced in P1.5 pituitary tissue in comparison to adult tissue. (G) The distribution of cell pairs in P1.5 tissue is compared to the adult cellular distribution. (H) Correlation vs distance plots show no large differences between randomised and non-randomised data over short cell-to-cell distances. Correlation vs distance plots are shown as described in Figure 3B. Images shown are maximum intensity projections of zstacks (E18.5: 165 µm; P1.5: 196 µm). Bar in images represents 100 µm. E18.5 (181 cells) and P1.5 (217 cells) data shown were taken from two independent representative experiments. hPRL, human prolactin.
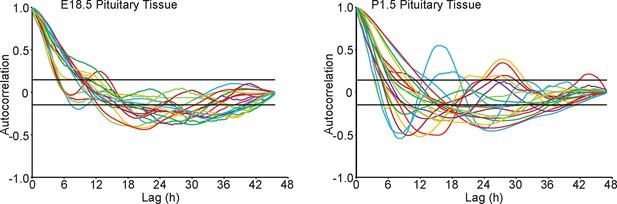
Autocorrelation analysis of fluorescence profiles from developing pituitary tissue.
Autocorrelation analysis of d2EGFP fluorescence intensity from cells shown in Figure 6B and 6F, with the 95% confidence interval representing a white noise process shown between black lines.
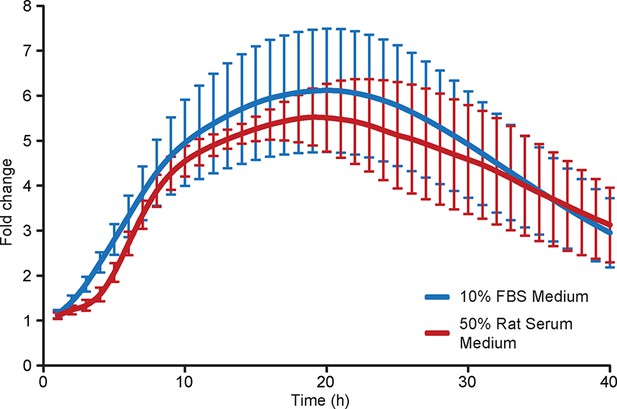
Comparison of prolactin transcription activity in adult pituitary tissue in different medium.
Graph showing the population transcriptional response from cells within adult pituitary tissue slices cultured in either 10% FBS or 50% rat serum supplemented medium in a closed system showing that differences in transcription profiles between immature and mature pituitaries was not due to the different culture media used. Prl-Luc49 rats were used and photon counts were collected using a photomultiplier tube (PMT) system. Data are from three independent pituitary cultures from two different animals (n = 6) for each condition, with the mean and SD shown. SD, standard deviation.
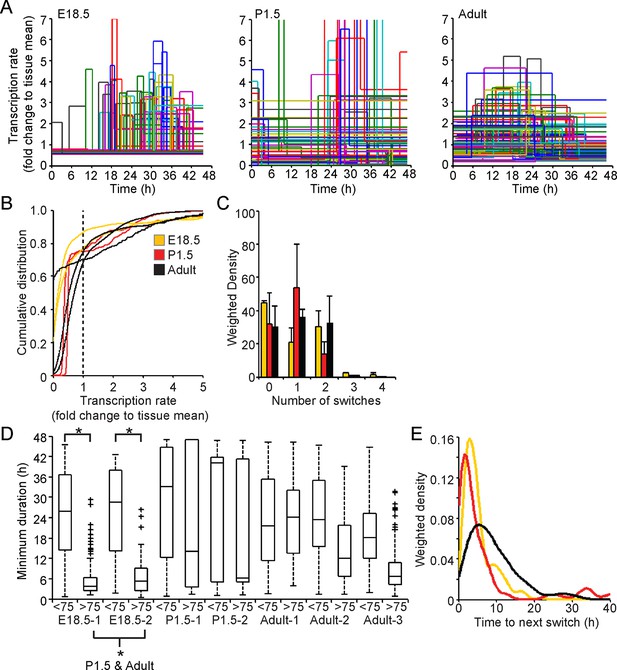
Comparison of prolactin transcription dynamics in different pituitary states.
(A) The stochastic switch model enables comparison of transcription dynamics in different pituitary states. Example plots of transcription rates estimated using the stochastic switch model. Analyses of populations of cells are shown from different states of pituitary tissue development. Each cell is associated with a single transcriptional profile obtained by random sampling of all the possible profiles for that cell (shown as in Figure 2A iii, with transcription rate and switch timings depicted). Data shown were from a single experiment representative of independent experiments (i: E18.5 n = 181 cells, ii: P1.5 n = 217 cells, iii: Adult n = 305 cells). (B–E) Characterisation of transcription dynamics estimated using the stochastic switch model. (B) The cumulative distribution function of the frequency of different transcription rates was estimated using a weighted kernel. No clear difference between pituitary developmental states was evident. (C) Weighted histogram of the number of switches for each developmental state. Data, pooled from independent experiments, show the mean + SD, calculated by weighting the number of switches in each profile by the probability of occurrence. P1.5 has significantly fewer switches to Adult and E18.5 (p<0.01 Mann-Whitney U test of the pooled distributions). (D) The duration of transcription rates for each tissue state. Transcription rates were binned into quartiles, with the first three lower quartiles grouped together (<75) as one bin and the highest quartile as the other bin (>75). The duration of transcriptional states was calculated for each bin. The duration represents the minimum duration spent in each transcriptional state, as not all transcriptional states were completely observed within the time-frame of the imaging experiment. Both E18.5 datasets showed a significant difference in the duration spent in low transcriptional states (lower 3 quartiles) to high transcriptional states (upper quartile) as did one P1.5 dataset and two Adult datasets. Moreover, the duration spent in the upper quartile of the E18.5 (pooled data) was significantly different to the duration spent in the upper quartile of either the pooled data of P1.5 or Adult. Significance was assessed through the distribution of the p-values calculated from a Mann-Whitney bootstrap (Figure 7—figure supplement 2). Boxplots were calculated by a random sampling of the weighted transcription rate distributions and the associated durations. (E) The graph shows a kernel density estimate of the time between any two switches using data from panel D, for cells that displayed more than one switch so that the total duration spent in a single transcriptional state was estimated. The inter-switch time in the adult tissues is longer than the inter switch times of either the E18.5 or P1.5 tissues. E18.5 = orange, P1.5 = red, Adult = black. All boxplots represent the median and interquartile range (IQR), with whiskers drawn 1.5xIQR away from the lower and upper quartile. SD, standard deviation.
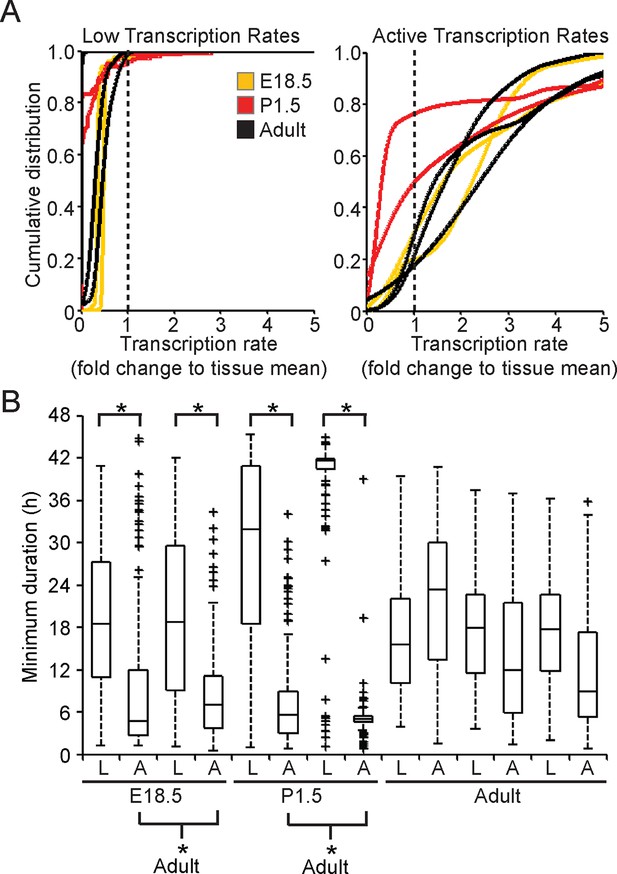
Characterisation of prolactin transcription dynamics.
(A) Characterisation of transcription rates, determined using the stochastic switch model, in E18.5, P1.5, and adult pituitaries. Rates were classified as ‘Low’ if they were preceded and followed by higher rates; all other rates were classified as ‘Active’ even if they were preceded by a higher level of activity, given that they were always followed by a further down-switch. Cells with no switches were removed from the analysis. Graphs are of the kernel density estimate of the cumulative distribution of transcription rates and show that low transcription rates were reduced compared to the active transcription rates. There is some evidence of reduced transcriptional rates in the ‘active’ states (lower rates relative to fold change of overall tissue) in the P1.5 datasets, but no clear difference in the distribution of rates employed by cells can be seen between Adult and E18.5. (B) Durations spent at 'low' (L) and 'active' (A) transcription rates. Boxplots of the duration spent in either an 'active' or 'low' transcriptional state. These are obtained by sampling from the weighted density of transcriptional states and calculating the associated duration. These show that 'active' transcription in E18.5 and P1.5 pituitaries occurs over shorter time periods than in adult pituitaries, as calculated through bootstrap Mann-Whitney U-tests. All boxplots represent the median and interquartile range (IQR), with whiskers drawn 1.5xIQR away from the lower and upper quartile.
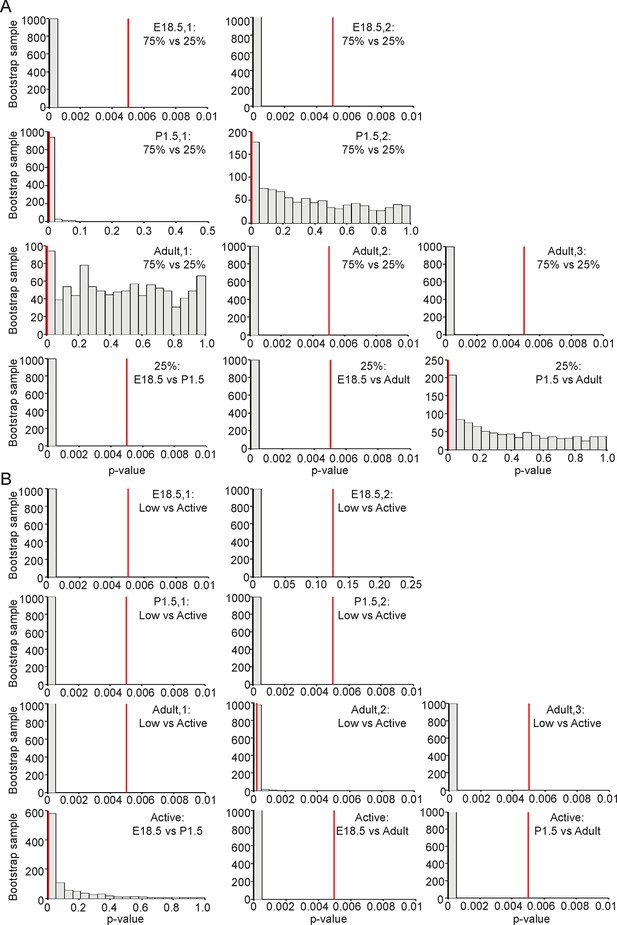
Significance testing of transcriptional state durations.
The boxplots in Figure 7D and Figure 7—figure supplement 1B are a single sample obtained from the weighted distribution of transcriptional rates. In order to test for significance difference between groups, bootstrap samples were obtained and a Mann-Whitney U-test was performed on each sample. The histograms of the p-values of each of these tests are shown in Figure 7—figure supplement 2A (corresponding to the tests on Figure 7D) and in Figure 7—figure supplement 2B (corresponding to the tests on Figure 7—figure supplement 1B). The vertical line indicates a p-value of 0.5% (an overall significance level of 5% with Bonferroni correction to account for multiple testing). Significance is indicated when the empirical histogram of the sampled p-values lies to the left of the red line. (A) Significant differences were found when comparing the duration spent in the lower transcriptional rates (lower three quartiles) or higher transcriptional rates (upper quartile) for datasets; Adult-2, Adult-3, E18.5-1 and E18.5-2. Significant differences were also found between the duration spent in the high transcriptional states of pooled datasets for E18.5 vs Adult and E18.5 vs P1.5. (B) Significant differences were found when comparing the duration spent in low (L) to active (A) transcriptional rates for datasets; Adult-1, Adult-3, P1.5-1, P1.5-2, E18.5-1 and E18.5-2. In addition, there is a significant difference in the duration spent in the high transcriptional states of the pooled datasets for E18.5 vs Adult and P1.5 vs Adult.
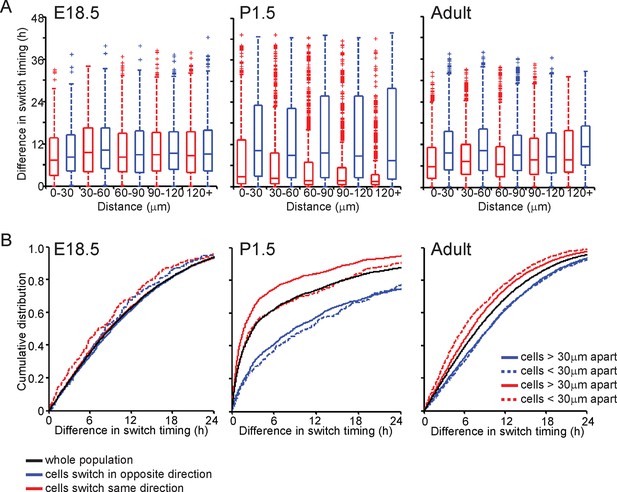
Spatial organisation of transcription switch profiles in developing pituitaries.
The spatial organisation of transcriptional switch profiles determined by the stochastic switch model in developing pituitaries was performed as outlined in Figure 4. (A) Graph of boxplots of switch timing intervals in cells that switch in the same direction and cells that switch in different directions, binned by the distance between cells. All pairwise switches are considered. An increase in the time interval between switches in transcription, which occur in the same direction, is seen with increasing distance between the cells in adult pituitary tissues but not in E18.5 or P1.5 pituitary tissues. (B) The cumulative distribution of the time interval between switch events. In adult pituitary tissue, cells within 30 µm that switch activity in the same direction do so within a smaller time frame than cells located greater than 30 µm apart, the unsorted population and cells that switch activity in opposite directions (confirmed by significant p-value <0.01 of Kolmogorov-Smirnov tests). In E18.5 and P1.5 pituitary tissues, this trend is not seen, as cells within 30 µm that switch activity in the same direction do so within a similar time frame to the whole cell population. In pooled data from P1.5 tissues, a significant difference (p-value <0.01 of Kolmogorov-Smirnov tests) is seen in the distribution of time intervals between cells that switch transcription activity in the same direction that are less than 30 µm apart and those that are greater than 30 µm apart, but this was not consistent across all data sets. All boxplots represent the median and interquartile range (IQR), with whiskers drawn 1.5xIQR away from the lower and upper quartile.
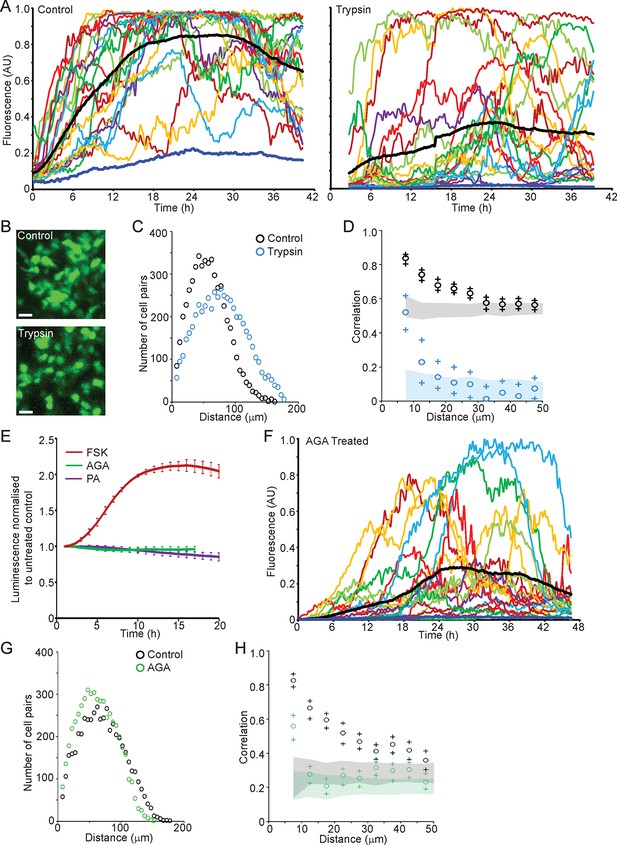
Cell communication influences the spatial organisation of prolactin transcription dynamics.
(A) Comparison of fluorescence profiles of hPRL-d2EGFP reporter gene activity from individual cells in control and trypsin-treated tissue. Cells in trypsin-treated tissue appeared less synchronised over time, but still showed an overall rise in activity as shown by the mean activity (black). The level of background fluorescence is shown in dark blue (mean from five areas). (B–D) Spatial correlation between fluorescence profiles of hPRL transcription activity is reduced in trypsin-treated tissue. (B) Images of cells within control and trypsin-treated tissue show that the distribution of cells and contacts between d2EGFP-expressing cells appeared altered following trypsin treatment. Bar represents 100 µm. (C) The intercellular distance between cells from control and trypsin-treated tissue was calculated as the median distance over the fluorescence imaging time-course (shown in A). (D) Correlation vs distance analyses showed a reduction in the difference between non-randomised and randomised data in trypsin-treated tissue compared to control, indicating a reduction in the spatial influence on transcription. (E–H) Inhibitors of gap junction signalling were used to assess whether juxtacrine signalling is influential in coordinating PRL transcription activity. (E) Real-time luminescence activity from populations of cells in primary cultures show that gap junction inhibitors (18α-glycyrrhetinic acid, AGA, and palmitoleic acid, PA) had little effect on overall PRL gene expression. Forskolin (FSK) was used as a positive control. (F) Fluorescence profiles of single cells in AGA treated tissue. Data are represented as described in (A). Transcription activity increased during the time-course (mean activity, black), similarly to control tissue (A). (G) The intercellular distance between cells from control and AGA-treated tissue was calculated as the median distance over the fluorescence imaging time-course (shown in A,F). (H) Correlation vs distance analyses showed a reduction in the difference between non-randomised and randomised fluorescence profiles in AGA treated tissue compared to control tissue indicating a reduction in the spatial coordination of transcription. Correlation vs distance plots are shown as described in Figure 3B.
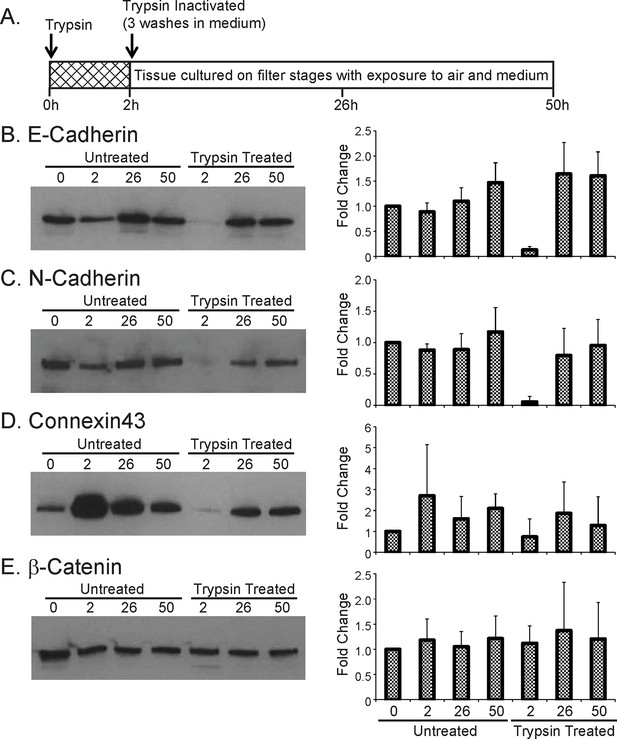
Effect of trypsin on cell junction proteins.
(A) Schematic of trypsin treatment regime. Tissues were incubated in trypsin, at 37°C for 2 hr without disaggregation and then cultured for defined time periods before analysis by western blot. (B) Representative western blot of E-Cadherin expression following trypsin treatment. The graph shows the quantification of E-Cadherin following trypsin treatment, expressed as the fold change from the initial untreated sample (0 hr), represented by the mean and SD. (C) Representative western blot of N-Cadherin expression following trypsin treatment. Protein expression levels were quantified as described in (B) and shown in the graph. (D) Representative western blot of Connexin43 expression following trypsin treatment. Protein expression levels were quantified as described in (B) and shown in the graph. (E) Representative western blot of β-Catenin expression following trypsin treatment. Protein expression levels were quantified as described in (B) and shown in the graph. SD, standard deviation.
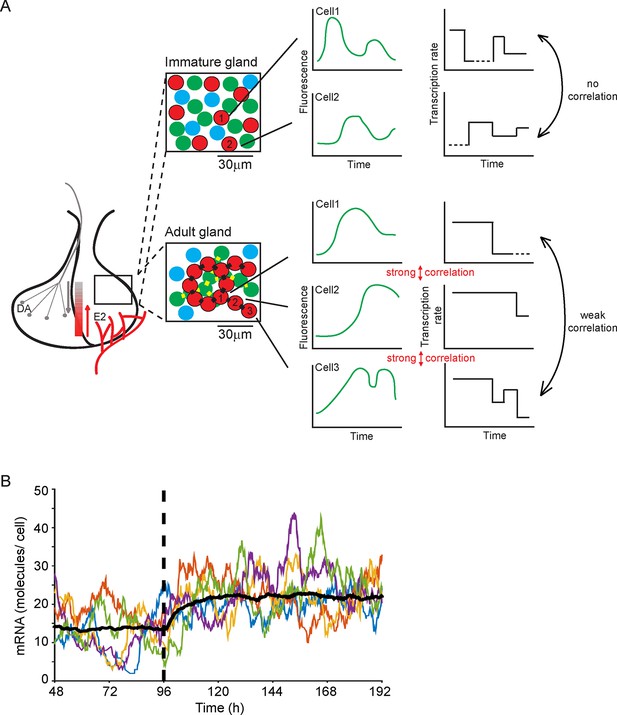
Multi-state transcription dynamics achieve robust tissue level responses.
(A) Organised cell networks and gap junction cell-cell signalling in adult pituitary glands (lower panels), but not in immature pituitary glands (upper panels), leads to local correlation of transcription between cells. Across a large population of cells transcription dynamics are essentially random and uncoordinated. Multi-state transcription dynamics ('off', 'primed' and 'on' at various levels) contribute to transcriptional heterogeneity between cells as the time spent in each state is exponentially distributed. The schematic indicates putative regional signalling and signalling gradients across the pituitary, for example from hypothalamic dopamine (DA) and systemic estradiol (E2). Segments of pituitary tissue are represented with lactotroph cells (red) connected via gap junctions (black rectangles). Examples of single cell transcription profiles (left plots) and their relation to multi-state transcription dynamics (right plots), with 'off', 'primed' (dashed lines) and different levels of transcription in the 'on' state are shown. (B) Simulation of the telegraph model with priming demonstrates how decreasing the 'primed” period (at the dashed line) gives rise to a rapid robust response across a population of cells showing heterogeneous activity. The simulation was performed on fluorescence profiles from 100 cells, with the mean of these responses shown (black line) along with 5 representative cell responses (coloured lines).





